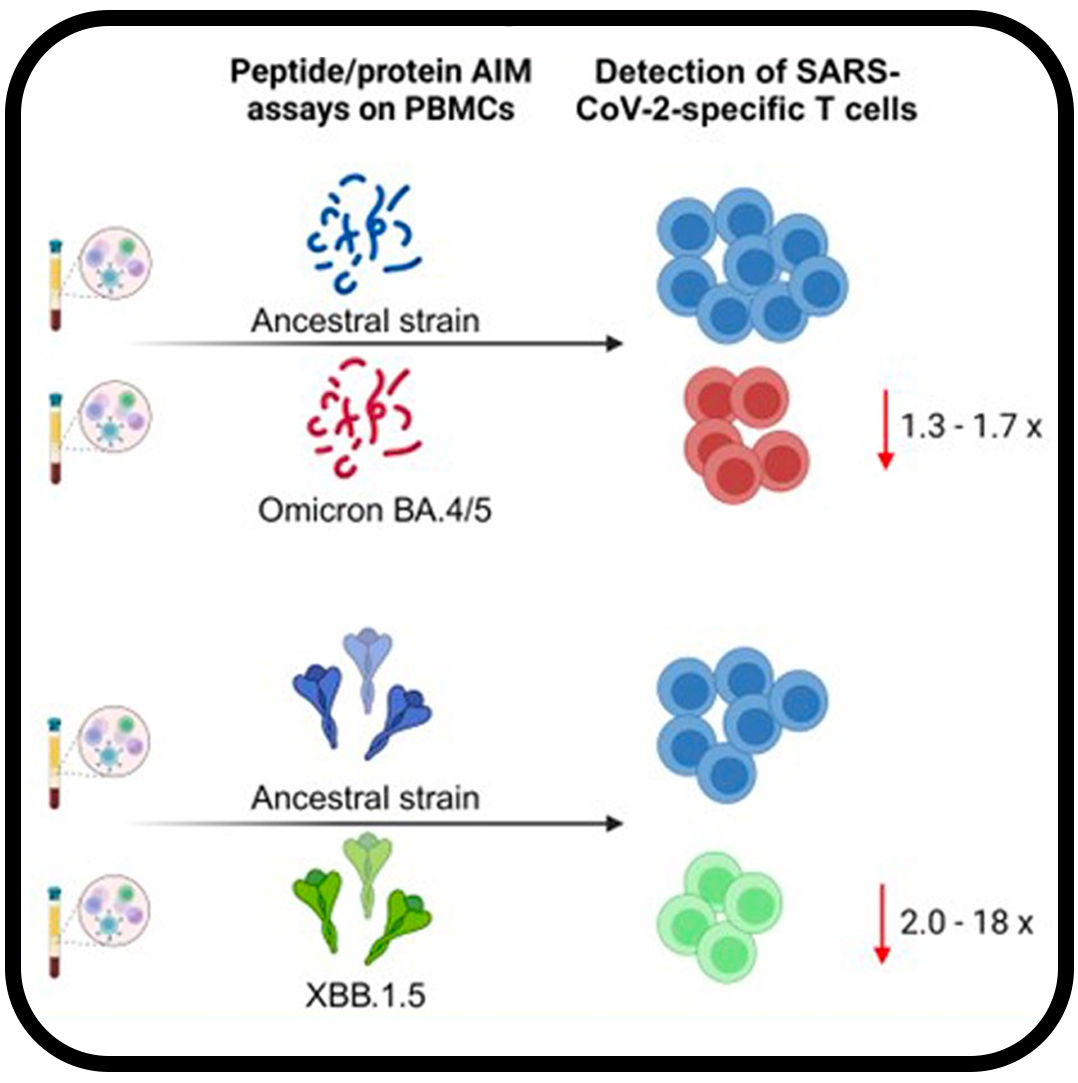
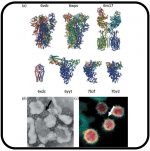
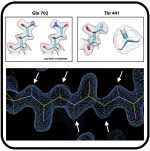
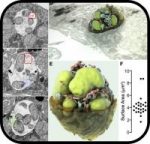
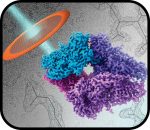
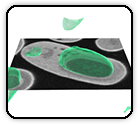
SARS-CoV-2 Variants Omicron BA.4/5 and XBB.1.5 Significantly Escape T Cell Recognition in Solid-organ Transplant Recipients Vaccinated Against the Ancestral Strain
Halvorson T, Ivison S, Huang Q, Ladua G, Yotis DM, Mannar D, Subramaniam S, Ferreira VH, Kumar D, Belga S, Levings MK; PREVenT Study Group.
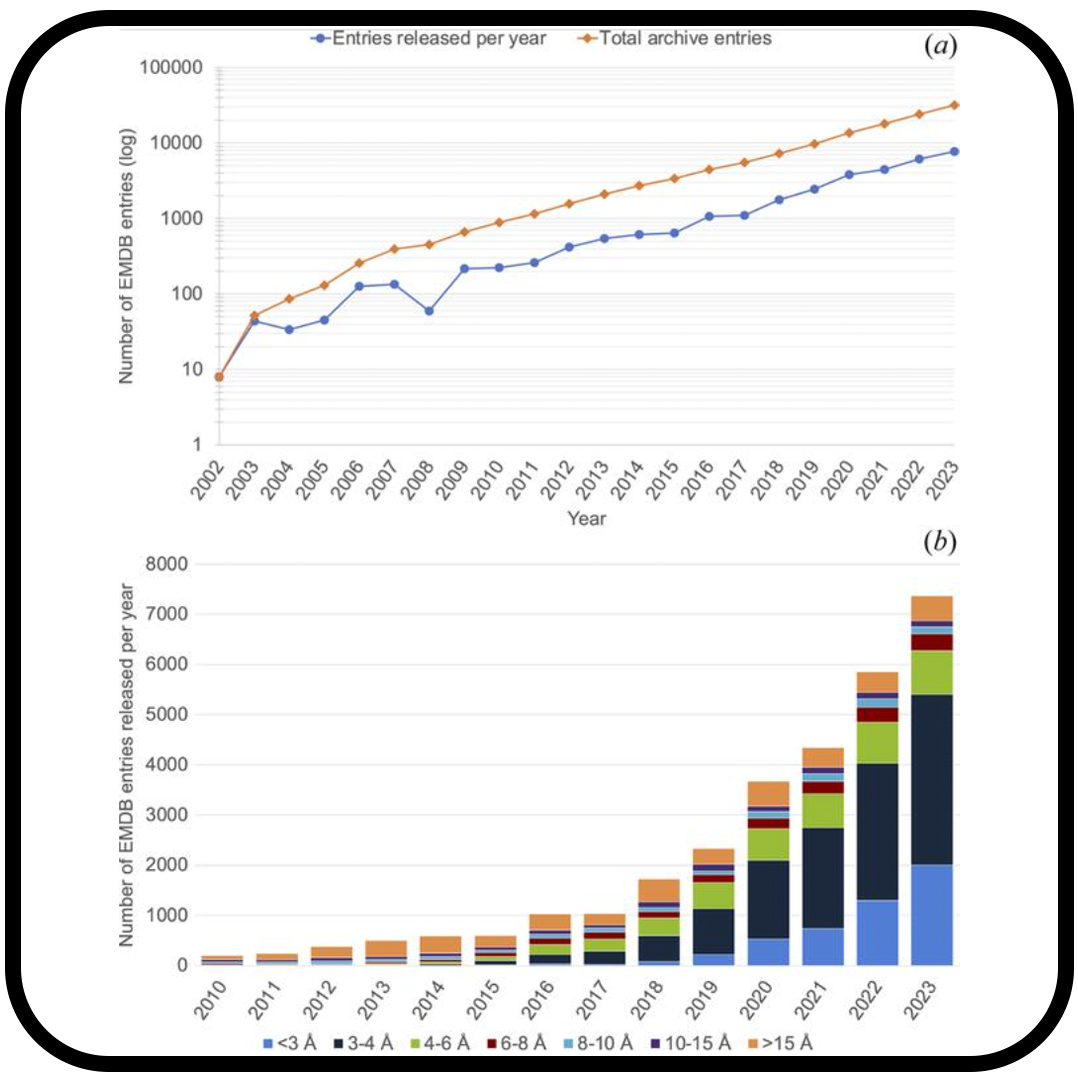
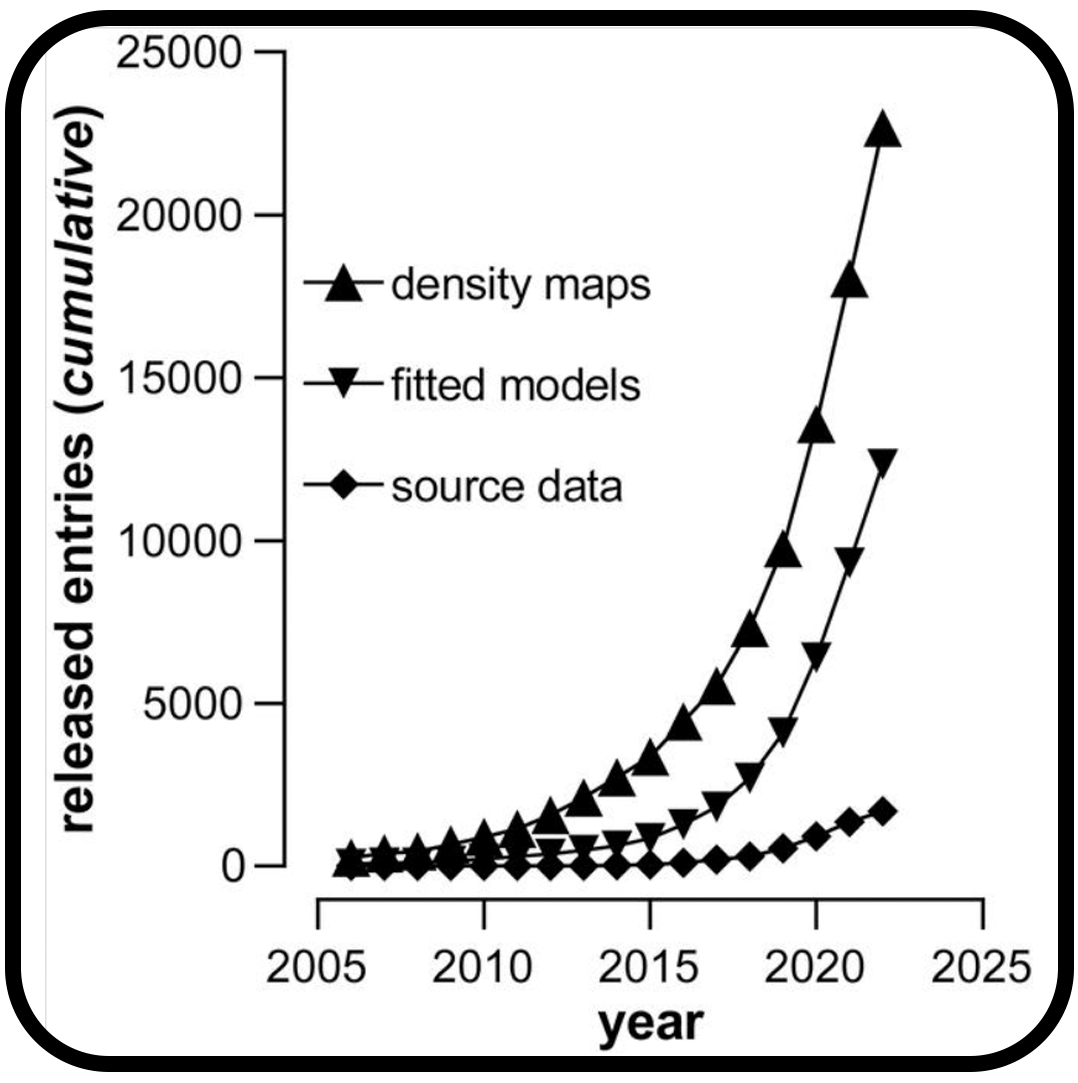
Community recommendations on cryoEM data archiving and validation
Kleywegt GJ, Adams PD, Butcher SJ, Lawson CL, Rohou A, Rosenthal PB, Subramaniam S, Topf M, Abbott S, Baldwin PR, Berrisford JM, Bricogne G, Choudhary P, Croll TI, Danev R, Ganesan SJ, Grant T, Gutmanas A, Henderson R, Heymann JB, Huiskonen JT, Istrate A, Kato T, Lander GC, Lok SM, Ludtke SJ, Murshudov GN, Pye R, Pintilie GD, Richardson JS, Sachse C, Salih O, Scheres SHW, Schroeder GF, Sorzano COS, Stagg SM, Wang Z, Warshamanage R, Westbrook JD, Winn MD, Young JY, Burley SK, Hoch JC, Kurisu G, Morris K, Patwardhan A, Velankar S.
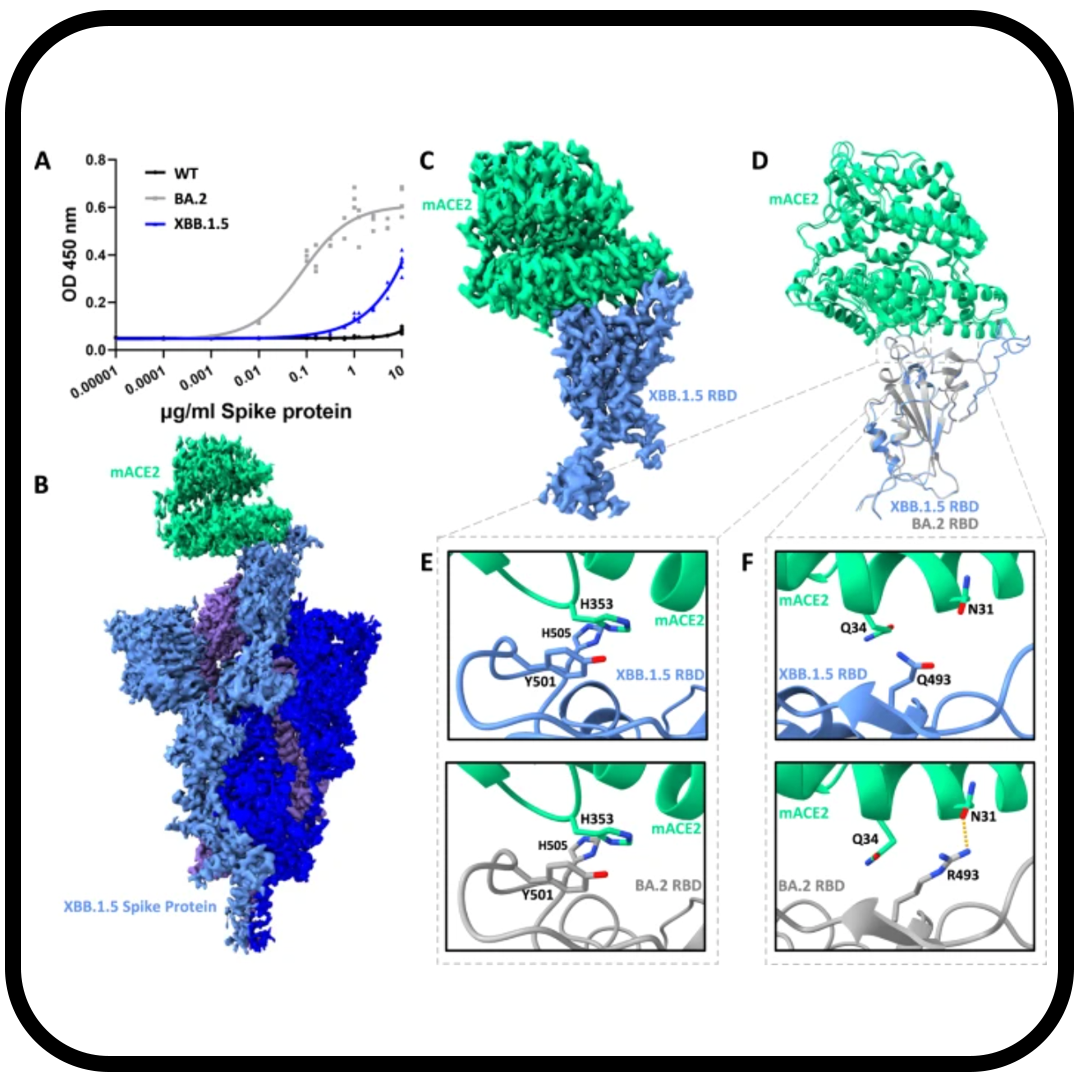
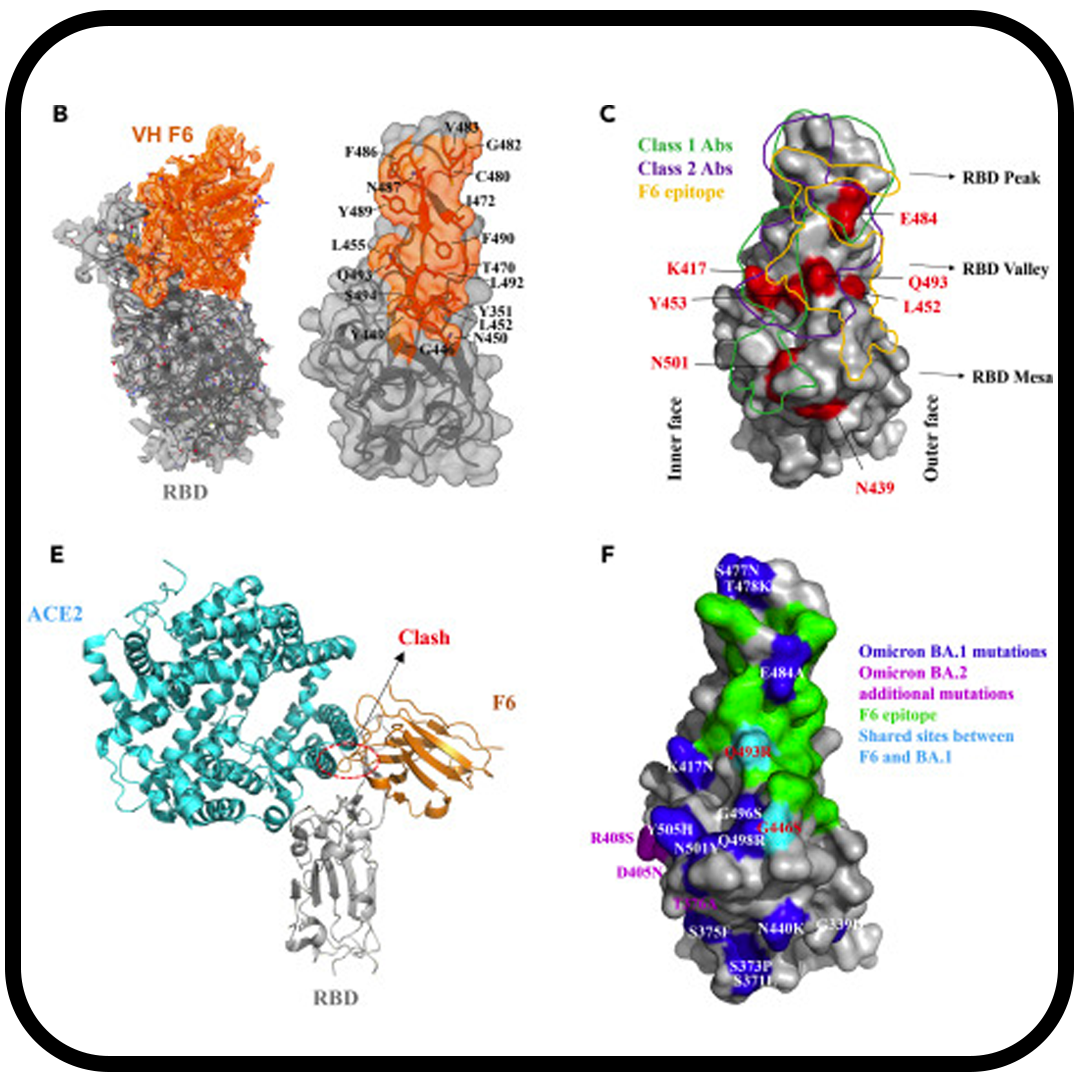
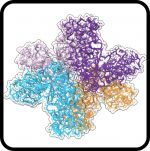
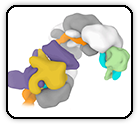
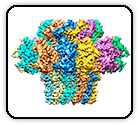
Altered receptor binding, antibody evasion and retention of T cell recognition by the SARS-CoV-2 XBB.1.5 spike protein
Mannar D, Saville JW, Poloni C, Zhu X, Berezuk A, Tidey K, Ahmed S, Tuttle KS, Vahdatihassani F, Cholak S, Cook L, Steiner TS, Subramaniam S.
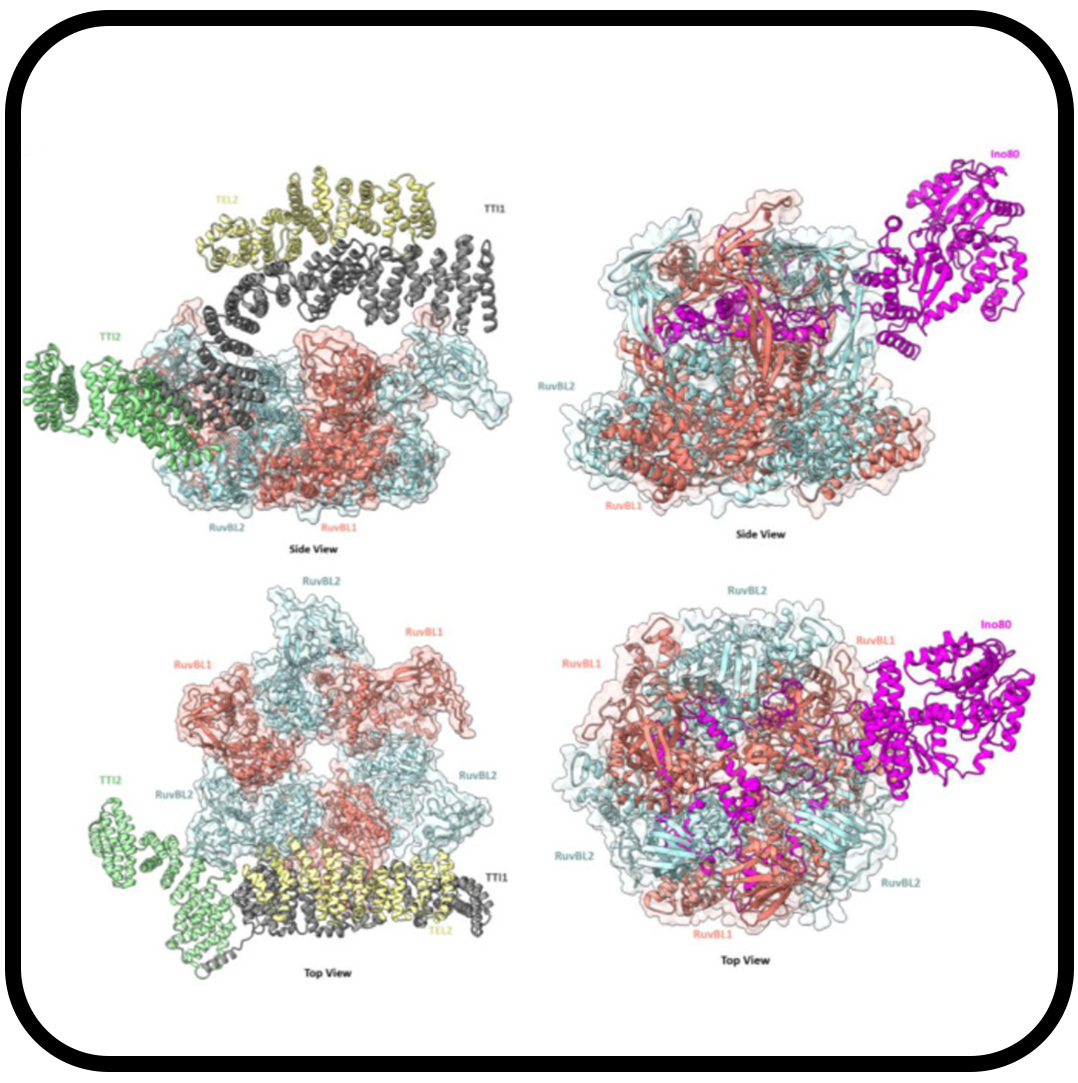
AAA ATPase protein-protein interactions as therapeutic targets in cancer
Mannar D, Ahmed S, Subramaniam S.
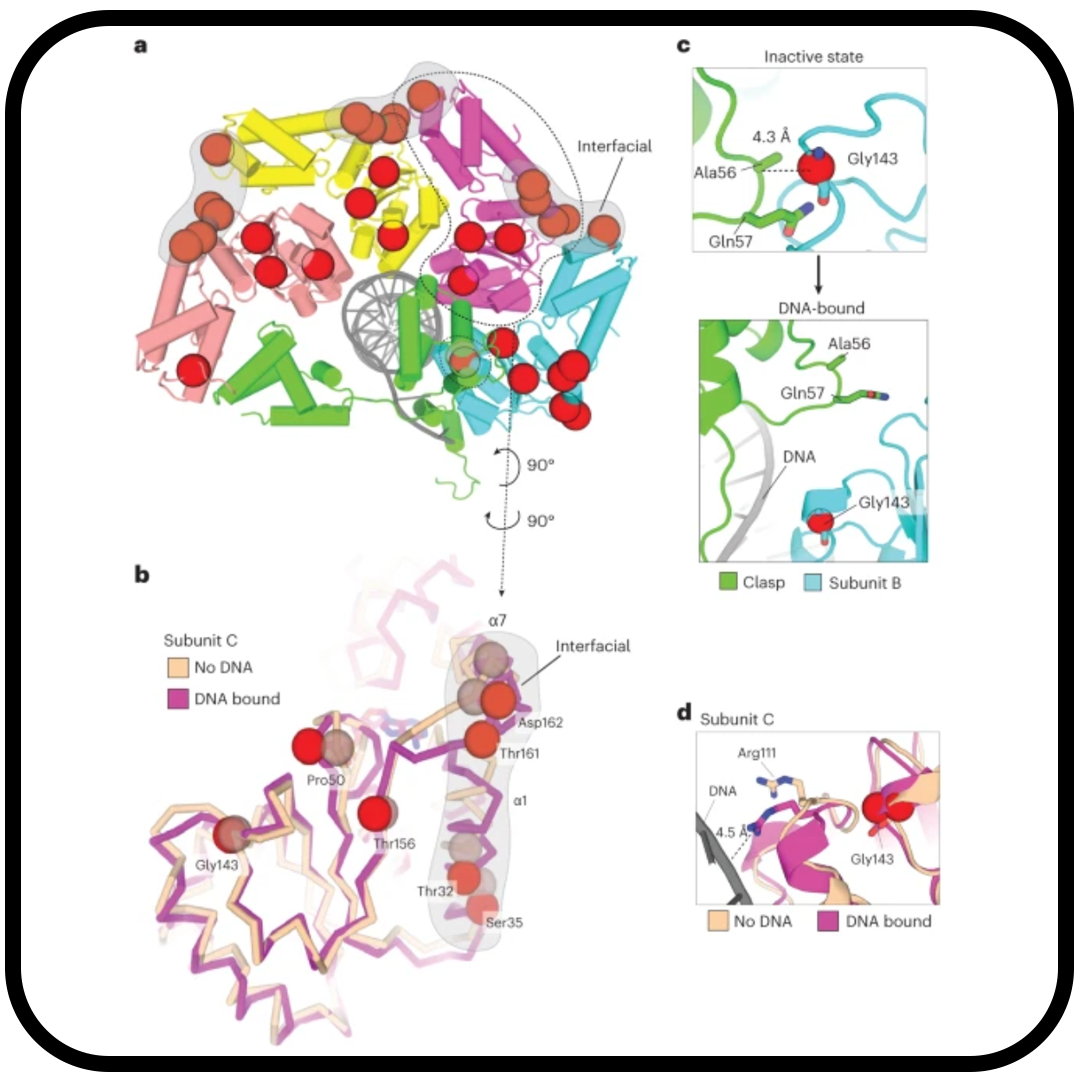
Autoinhibition of a clamp-loader ATPase revealed by deep mutagenesis and cryo-EM
Marcus K, Huang Y, Subramanian S, Gee CL, Gorday K, Ghaffari-Kashani S, Luo XR, Zheng L, O’Donnell M, Subramaniam S, Kuriyan J.
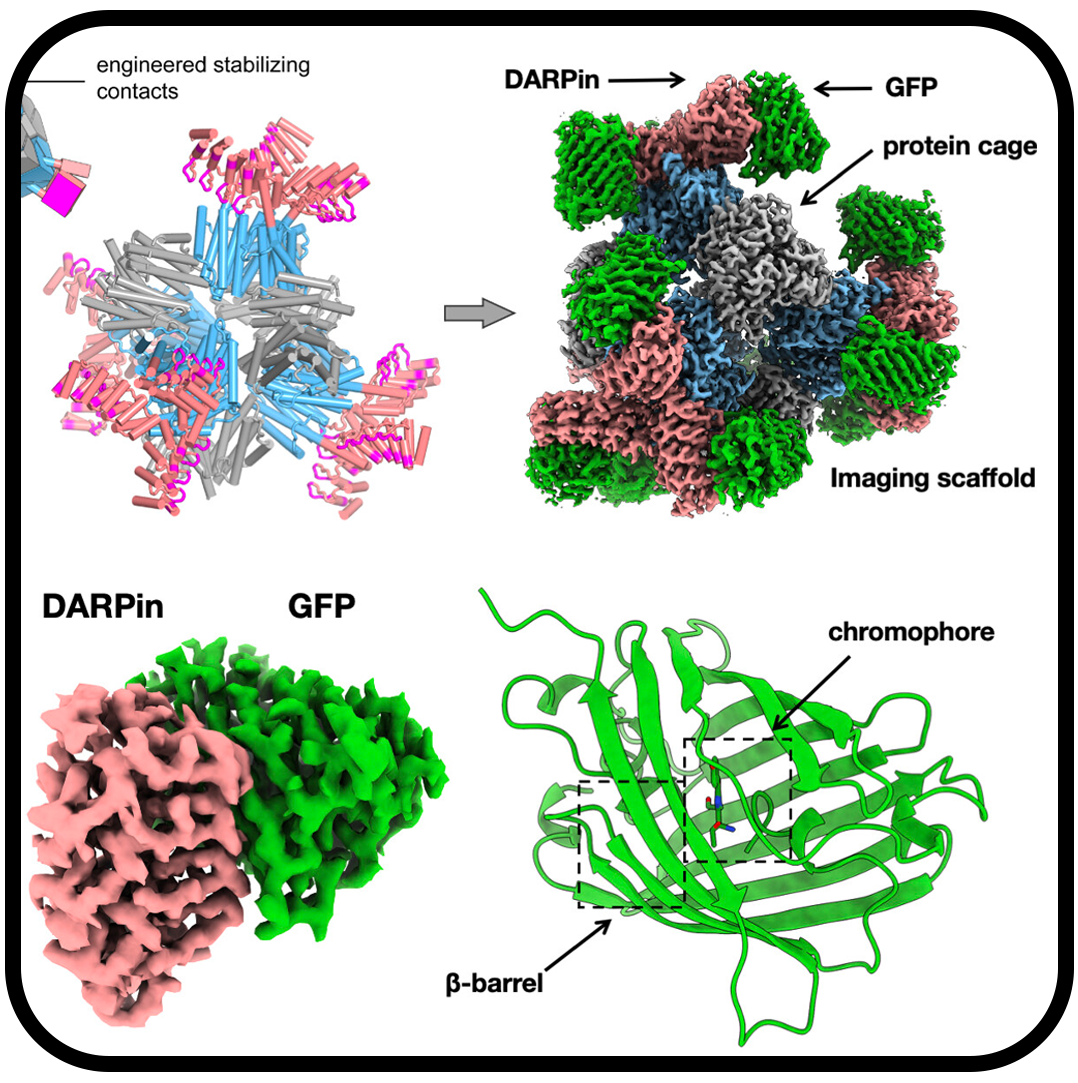
Cryo-EM structure determination of small therapeutic protein targets at 3 Å-resolution using a rigid imaging scaffold
Castells-Graells R, Meador K, Arbing MA, Sawaya MR, Gee M, Cascio D, Gleave E, Debreczeni JÉ, Breed J, Leopold K, Patel A, Jahagirdar D, Lyons B, Subramaniam S, Phillips C, Yeates TO.
Proceedings of the National Academy of Sciences USA 120: e2305494120 (2023)
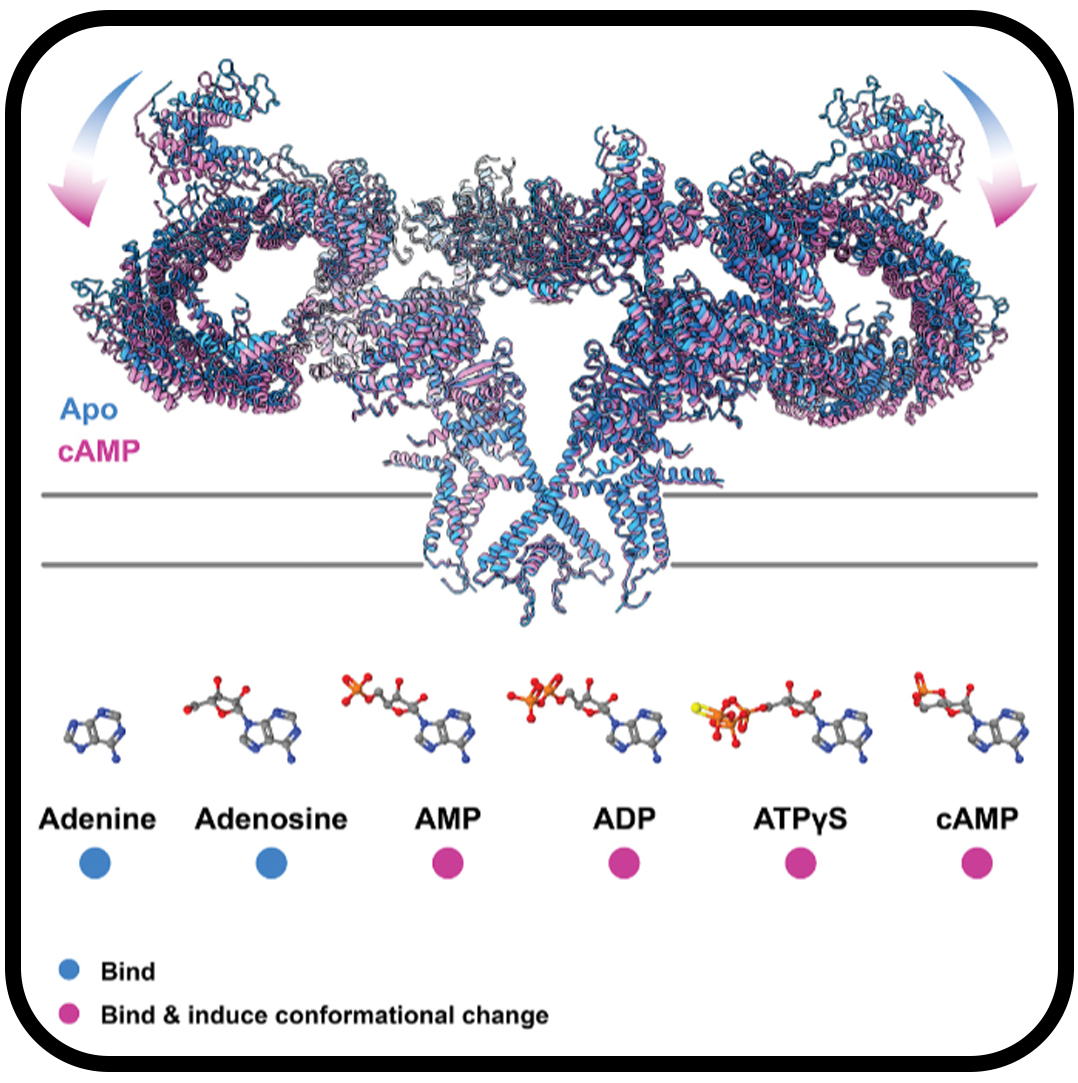
Allosteric modulation of ryanodine receptor RyR1 by nucleotide derivatives
Cholak S, Saville JW, Zhu X, Berezuk AM, Tuttle KS, Haji-Ghassemi O, Alvarado FJ, Van Petegem F, Subramaniam S.
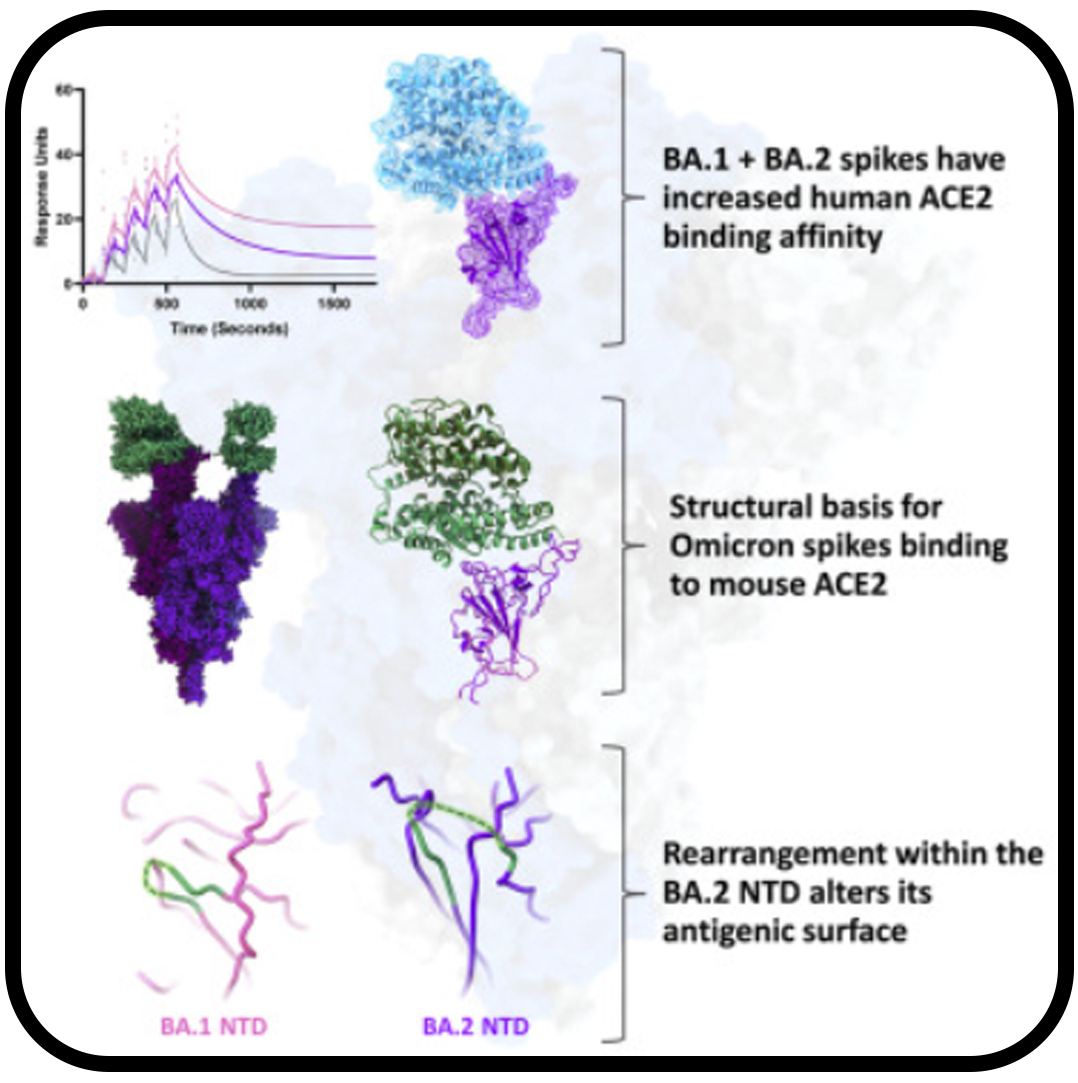
Structural analysis of receptor engagement and antigenic drift within the BA.2 spike protein
Saville JW, Mannar D, Zhu X, Berezuk AM, Cholak S, Tuttle KS, Vahdatihassani F, Subramaniam S.
Learning from Gobind Khorana: “The difficult we do today, the impossible tomorrow”
Subramaniam S.
Introduction: Cryo-EM in Biology and Materials Research
Subramaniam S, Danino D.
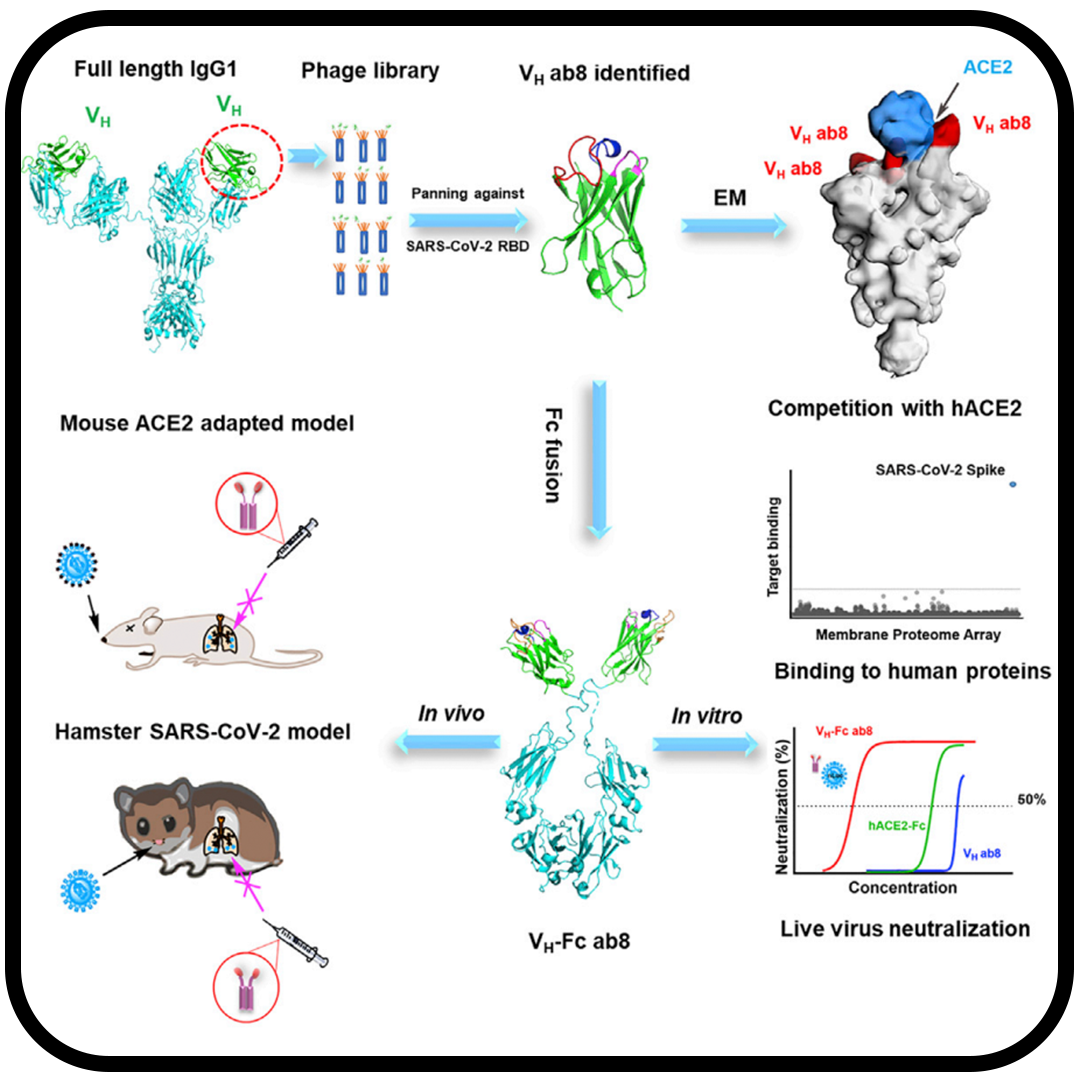
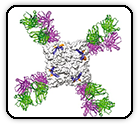
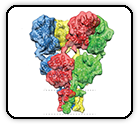
Potent and broad neutralization of SARS-CoV-2 variants of concern (VOCs) including omicron sub-lineages BA.1 and BA.2 by biparatopic human VH domains
Chen C, Saville JW, Marti MM, Schäfer A, Cheng MH, Mannar D, Zhu X, Berezuk AM, Banerjee A, Sobolewski MD, Kim A, Treat BR, Castanha PMDS, Enick N, McCormick KD, Liu X, Adams C, Hines MG, Sun Z, Chen W, Jacobs JL, Barratt-Boyes SM, Mellors JW, Baric RS, Bahar I, Dimitrov DS, Subramaniam S, Martinez DR, Li W.
Imaging Cellular Architecture in Three Dimensions Through Electron Microscopy
SARS-CoV-2 variants of concern: spike protein mutational analysis and epitope for broad neutralization
,,,, ,, ,, ,,,,, , ,, ,,
Three-Dimensional Visualization of Viral Structure, Entry, and Replication Underlying the Spread of SARS-CoV-2
Saville JW, Berezuk AM, Srivastava SS, Subramaniam S.
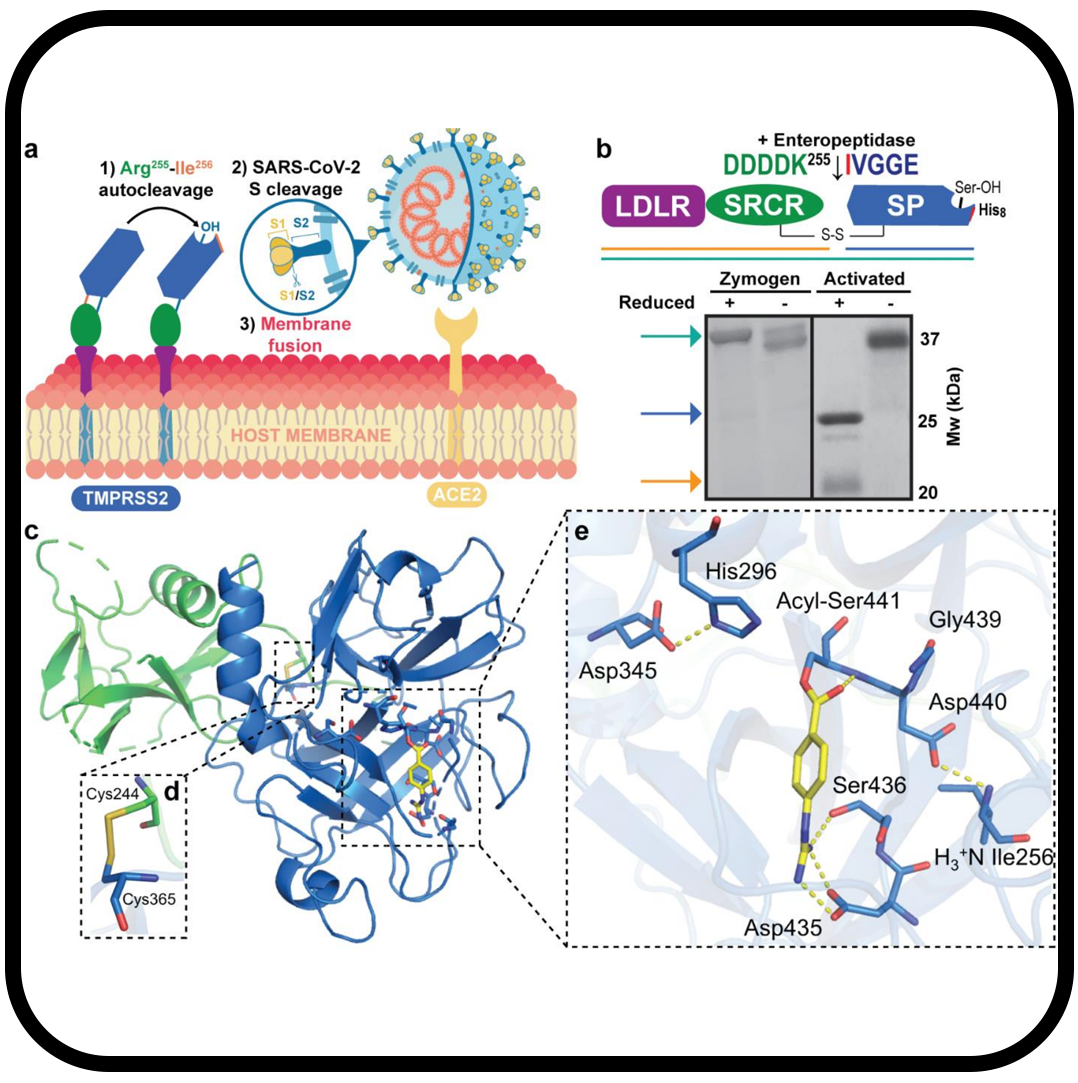
Structure and activity of human TMPRSS2 protease implicated in SARS-CoV-2 activation
Fraser BJ, Beldar S, Seitova A, Hutchinson A, Mannar D, Li Y, Kwon D, Tan R, Wilson RP, Leopold K, Subramaniam S, Halabelian L, Arrowsmith CH, Bénard F.
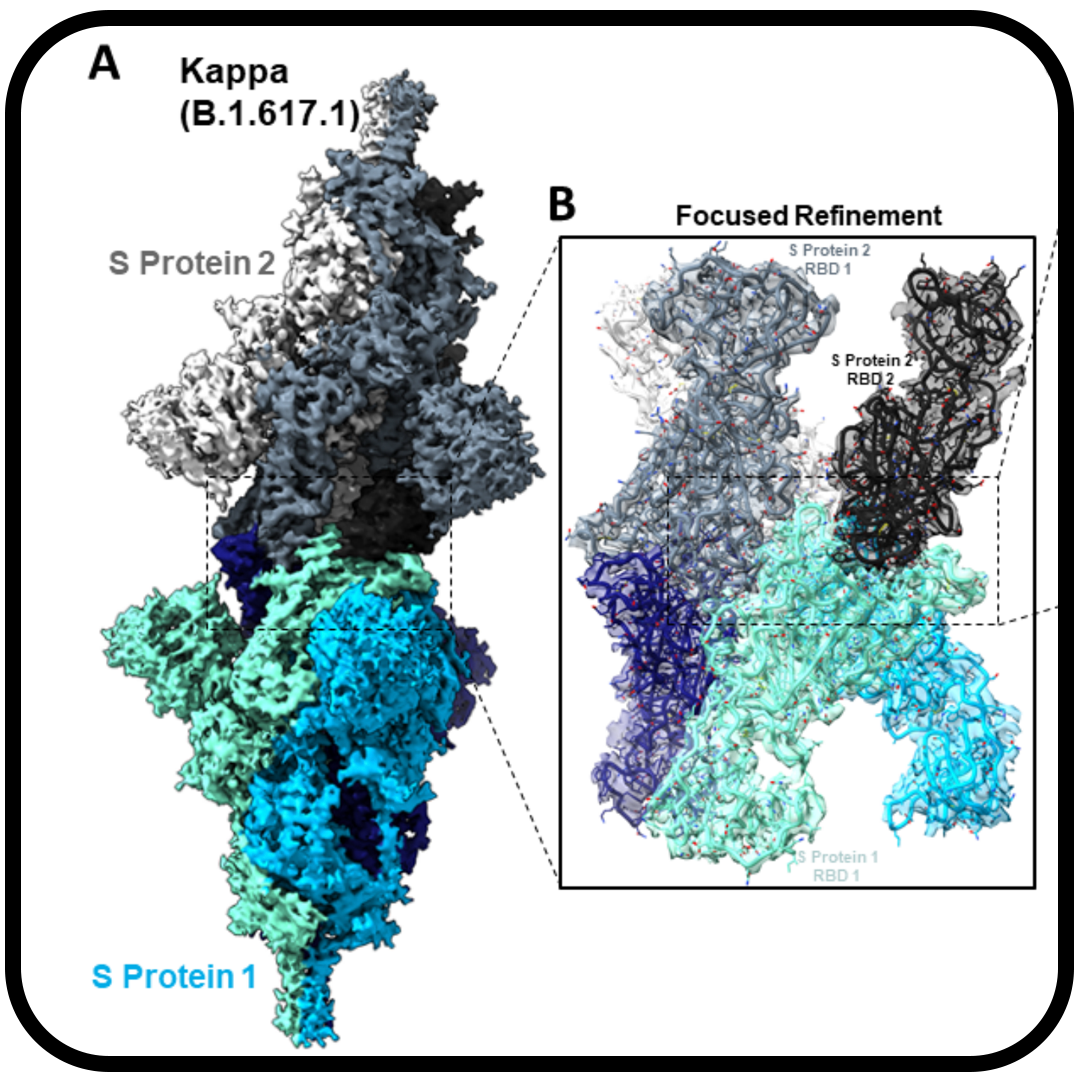
Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants
,,, ,,,,,,,, ,
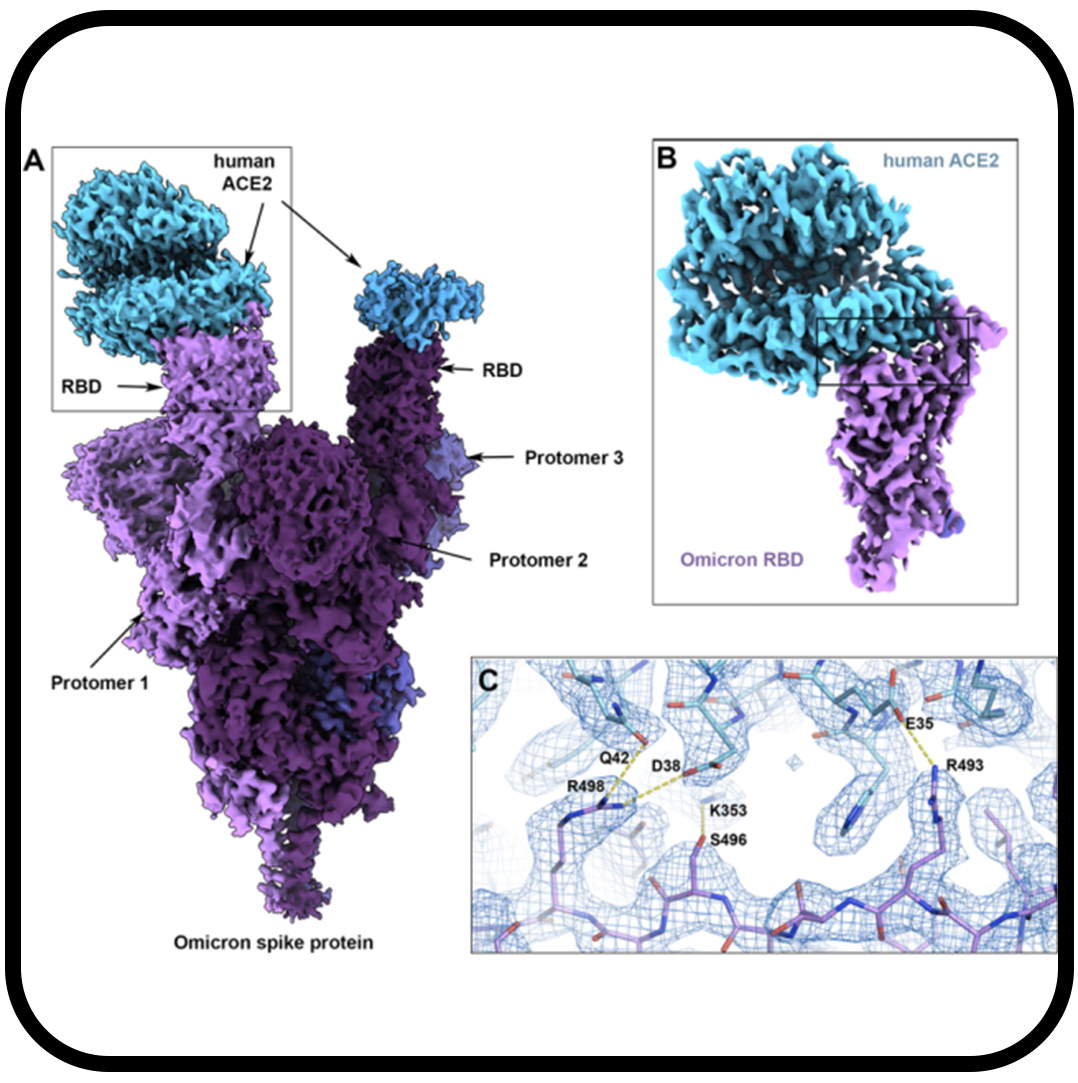
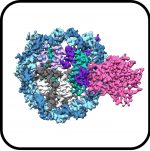
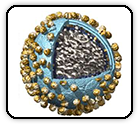
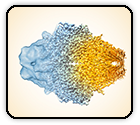
SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex
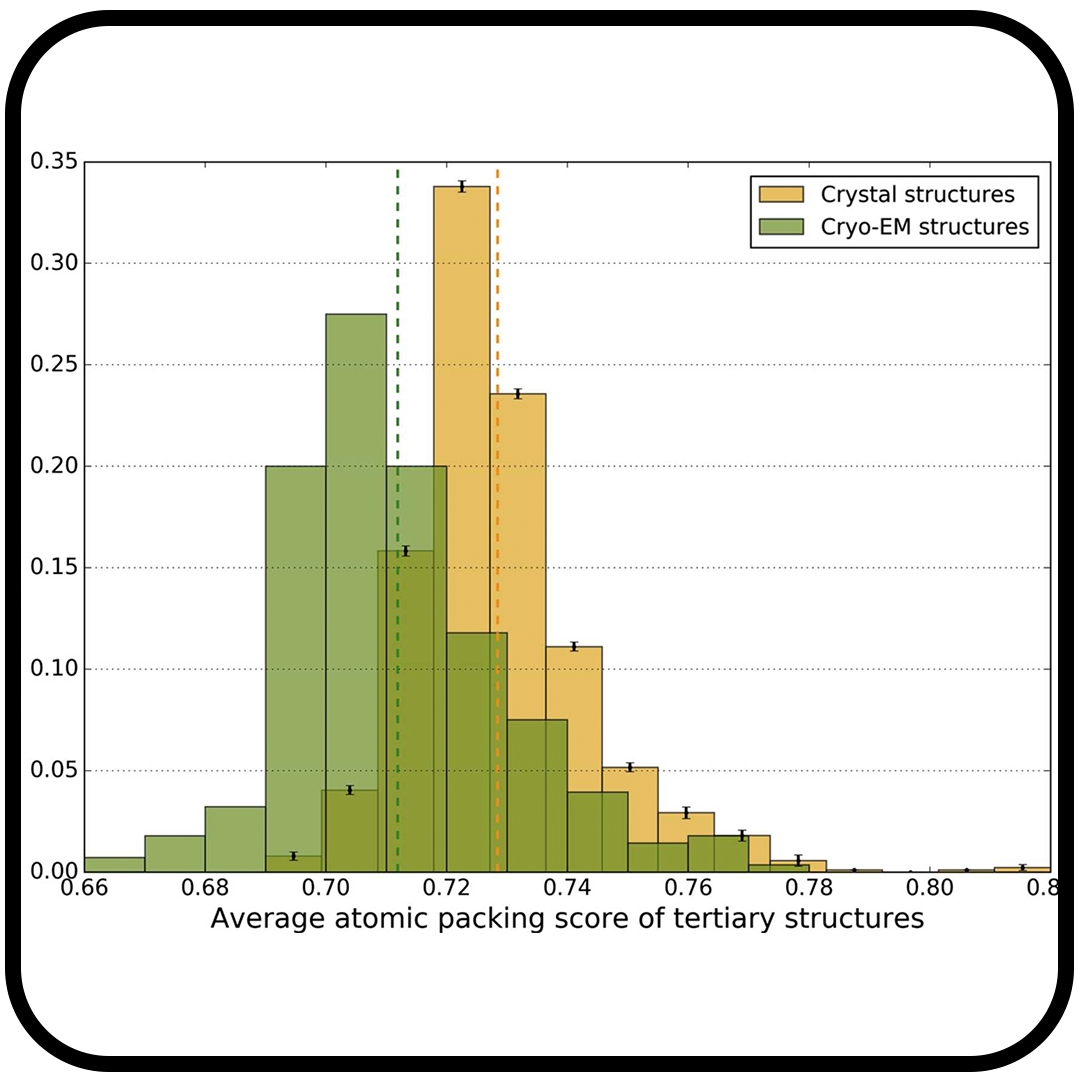
Comparison of side-chain dispersion in protein structures determined by cryo-EM and X-ray crystallography
Ravikumar A, Gopnarayan MN, Sriram Subramaniam S, Srinivasan N.
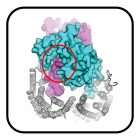
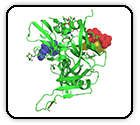
Structural Analysis of Receptor Binding Domain Mutations in SARS-CoV-2 Variants of Concern that Modulate ACE2 and Antibody Binding
Mannar D, Saville JW, Zhu X, Srivastava SS, Berezuk AM, Zhou S, Tuttle KS, Kim A, Li W, Dimitrov DS, Subramaniam S.
A molecular mechanism for the generation of ligand-dependent differential outputs by the epidermal growth factor receptor
Huang Y, Ognjenovic J, Karandur D, Miller K, Merk A, Subramaniam S, Kuriyan J.
AAA+ ATPase p97/VCP mutants and inhibitor binding disrupt inter-domain coupling and subsequent allosteric activation
Caffrey B, Zhu X, Berezuk A, Tuttle K, Chittori S, Subramaniam S.
New insights into Raf regulation from structural analyses
Kondo Y, Paul JW III, Subramaniam S, Kuriyan J.
Glycan reactive anti-HIV-1 antibodies bind the SARS-CoV-2 spike protein but do not block viral entry
Mannar D, Leopold K, Subramaniam S.
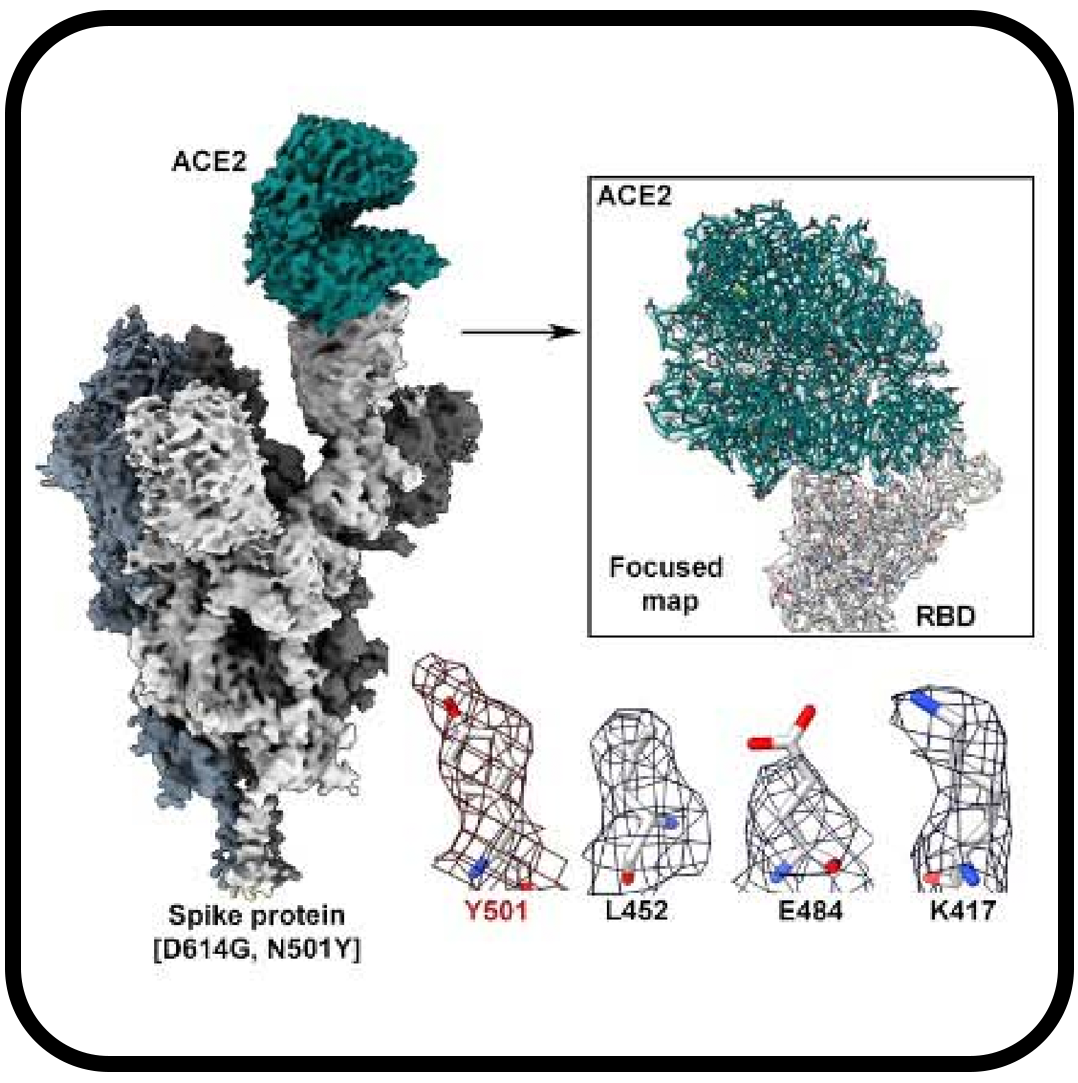
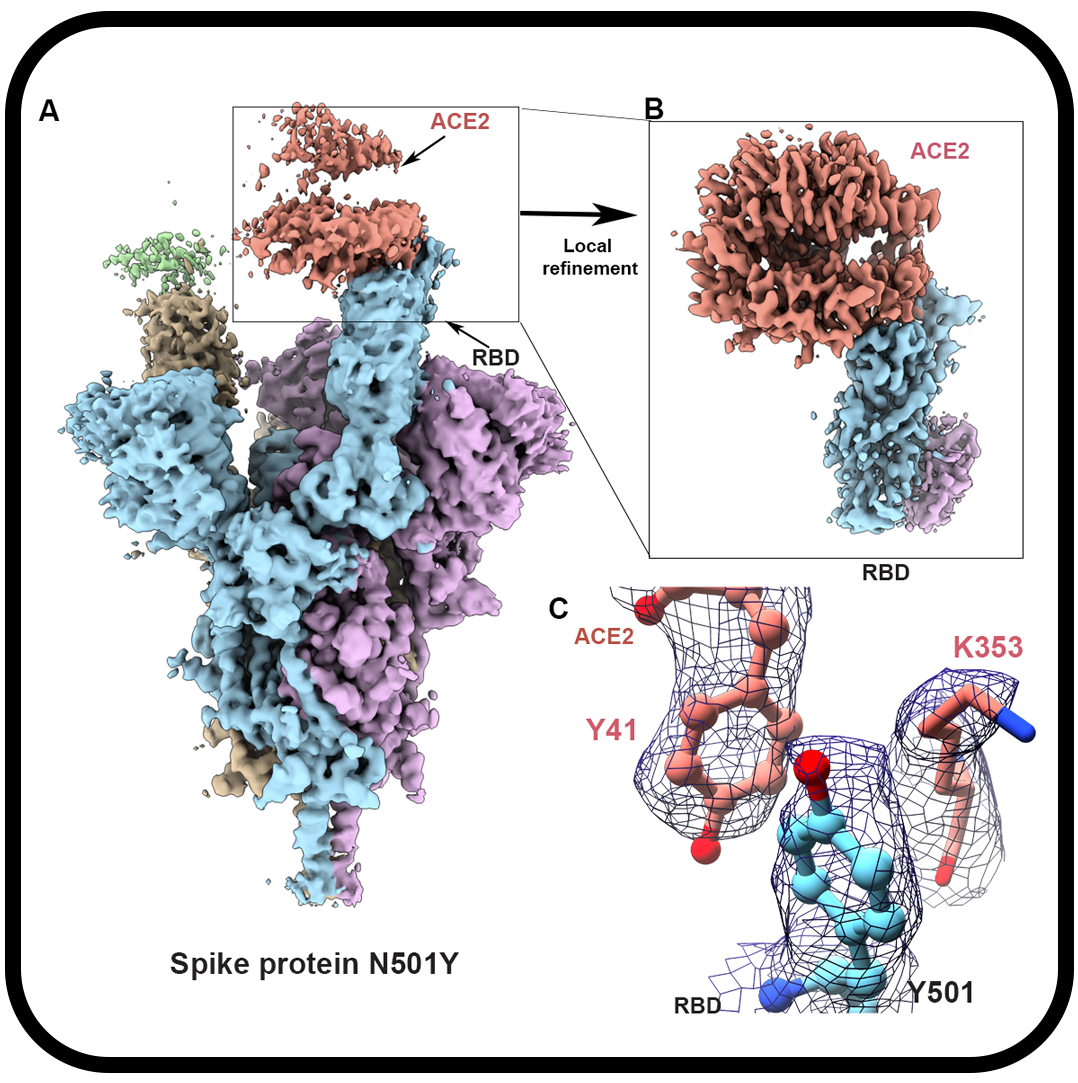
Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies
Zhu X, Mannar D, Srivastava SS, Berezuk AM, Demers JP, Saville JW, Leopold K, Li W, Dimitrov DS, Tuttle KS, Zhou S, Chittori S, Subramaniam S.
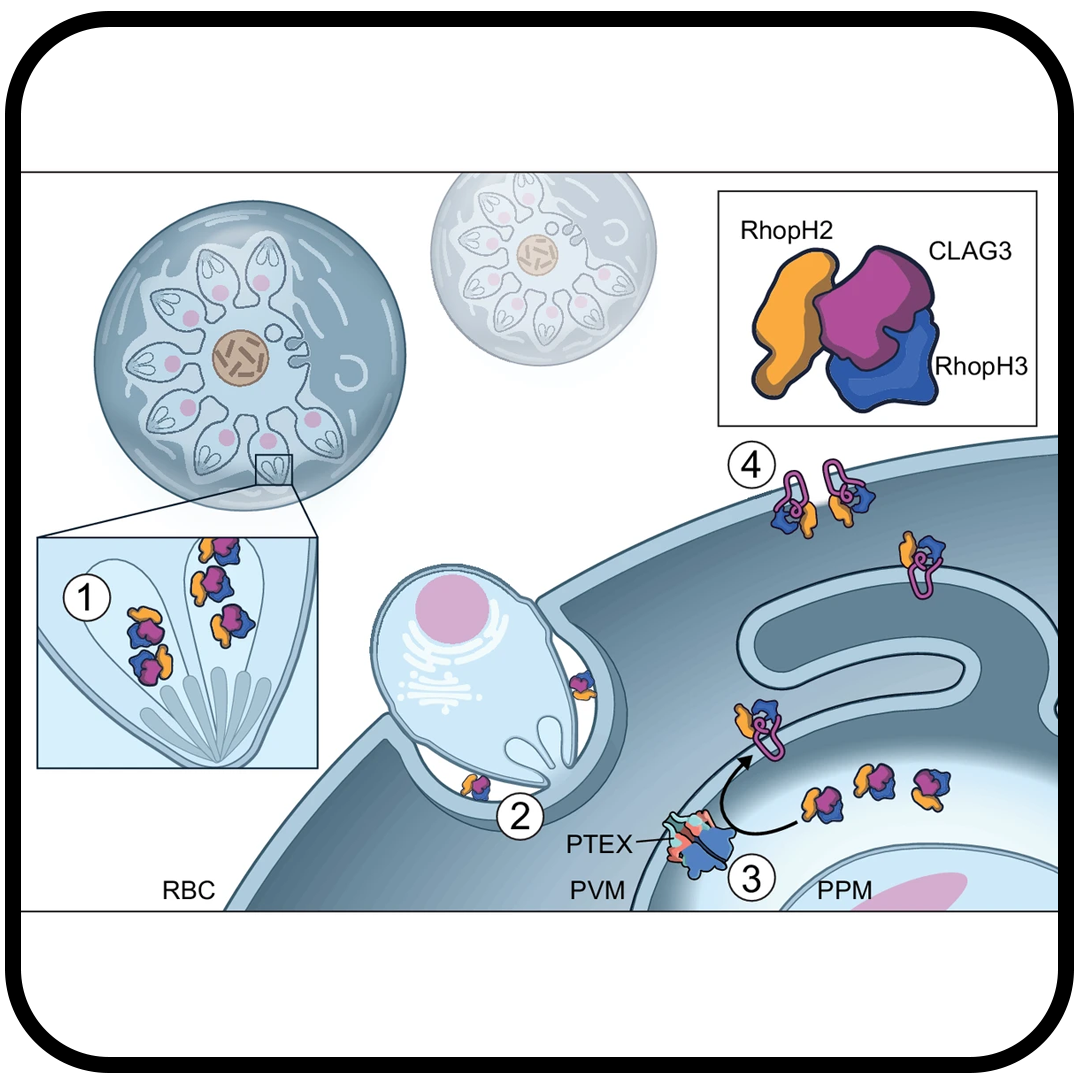
Malaria parasites use a soluble RhopH complex for erythrocyte invasion and an integral form for nutrient uptake
Schureck MA, Darling JE, Merk A, Shao J, Daggupati G, Srinivasan P, Olinares PDB, Rout MP, Chait BT, Wollenberg K, Subramaniam S, Desai SA.
Visualizing Cancer
Krummel M, Hu K, de Vries EGE, Goetz JG, Passaro D, Subramaniam S, Misteli T, Skala M, Bravo-Cordero JJ, Sutcliffe JL.
High Potency of a Bivalent Human Vh Domain in SARS-CoV-2 Animal Models
Li W, Schäfer A, Kulkarni SS, Liu X, Martinez DR, Chen C, Sun Z, Leist SR, Drelich A, Zhang L, Ura ML, Berezuk A, Chittori S, Leopold K, Mannar D, Srivastava SS, Zhu X, Peterson EC, Tseng CT, Mellors JW, Falzarano D, Subramaniam S, Baric RS, Dimitrov DS.
1.8 Å resolution structure of β-galactosidase with a 200 kV CRYO ARM electron microscope
Merk A, Fukumura T, Zhu X, Darling JE, Grisshammer R, Ognjenovic J, Subramaniam S.
The National Cryo-EM Facility at Frederick National Laboratory: Operational Procedures, Methods, and Outputs
Baxa U, Edwards TJ, Hutchison M, Wier AD, Finney J, Wang H, Subramaniam S.
Microscopy Today, Cambridge University Press 28: 12–17 (2020)
Host membrane lipids are trafficked to membranes of intravacuolar bacterium Ehrlichia chaffeensis
Lin M, Grandinetti G, Hartnell LM, Bliss D, Subramaniam S, Rikihisa Y.
Proceedings of the National Academy of Sciences USA 117: 8032-43 (2020)
Plasmodium vivax and human hexokinases share similar active sites but display distinct quaternary architectures
Srivastava SS, Darling JE, Suryadi J, Morris JC, Drew ME, Subramaniam S.
Biochemical and structural analyses reveal that the tumor suppressor neurofibromin (NF1) forms a high-affinity dimer
Sherekar M, Han SW, Ghirlando R, Messing S, Drew M, Rabara D, Waybright T, Juneja P, O’Neill H, Stanley CB, Bhowmik D, Ramanathan A, Subramaniam S, Nissley DV, Gillette W, McCormick F, Esposito D.
Cryo-electron microscopy instrumentation and techniques for life sciences and materials science
Williams R, McComb D, Subramaniam S.
Structure of the primed state of the ATPase domain of chromatin remodeling factor ISWI bound to the nucleosome
Chittori S, Hong J, Bai Y, Subramaniam S.
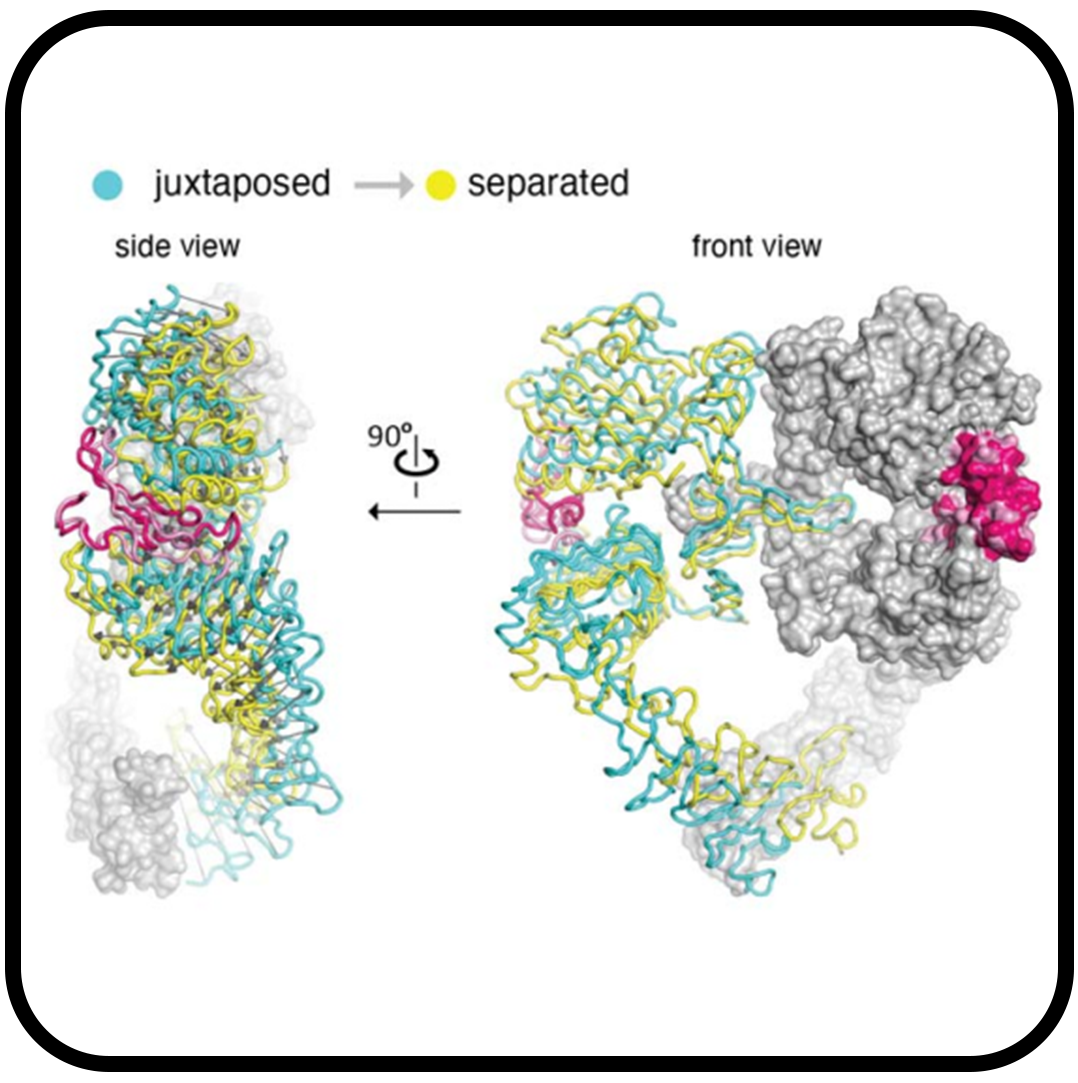
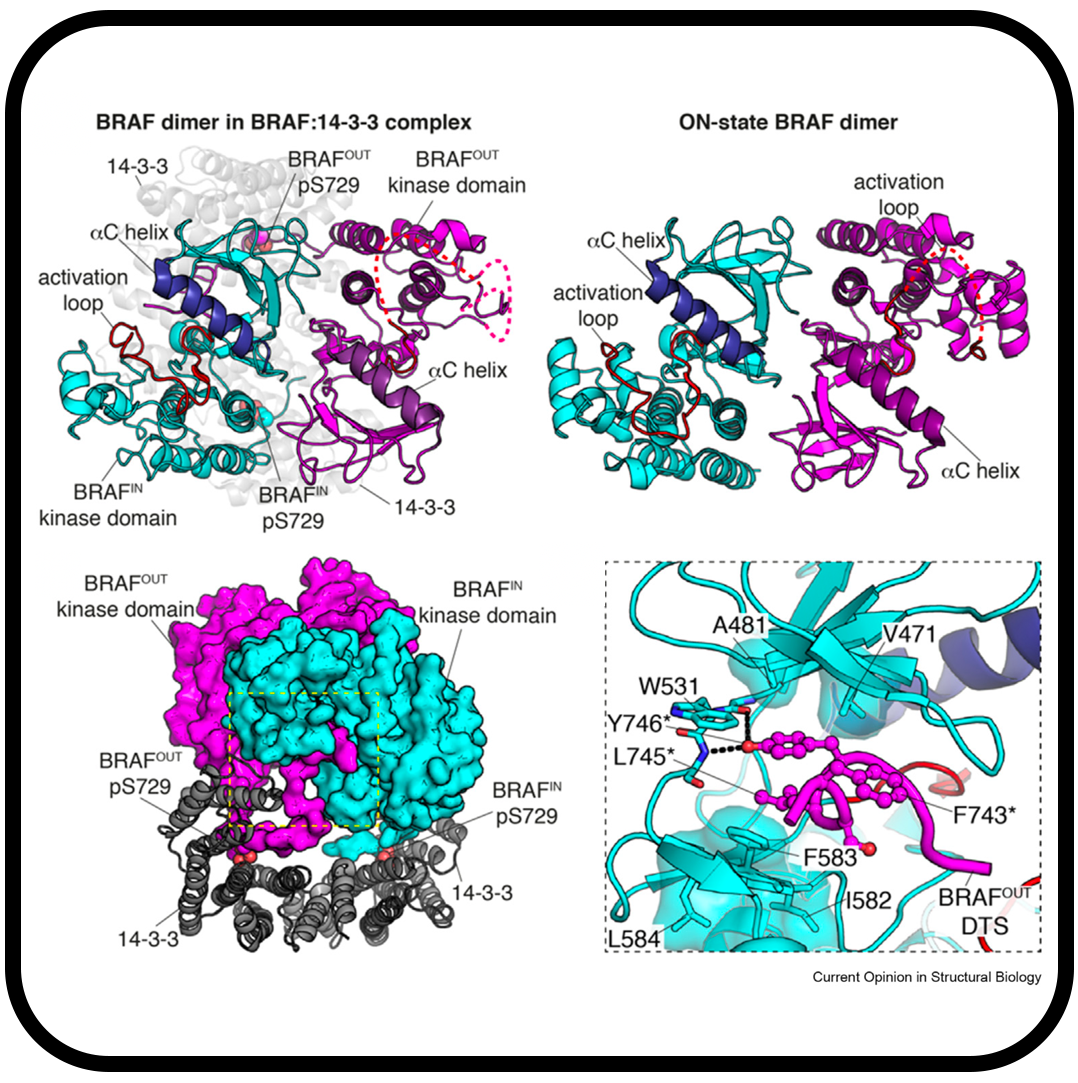
Cryo-EM structure of a dimeric B-Raf:14-3-3 complex reveals asymmetry in the active sites of B-Raf kinases
Kondo Y, Ognjenović J, Banerjee S, Karandur D, Merk A, Kulhanek K, Wong K, Roose JP, Subramaniam S, Kuriyan J.
Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9
Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonovic M, Subramaniam S.
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex
Marabelli C, Marrocco B, Pilotto S, Chittori S, Picaud S, Marchese S, Ciossani G, Forneris F, Filippakopoulos P, Schoehn G, Rhodes D, Subramaniam S, Mattevi A.
Semi-Automated 3D segmentation of human skeletal muscle using Focused Ion Beam-Scanning Electron Microscopic images
Caffrey BJ, Maltsev AV, Gonzalez-Freire M, Hartnell LM, Ferrucci L, Subramaniam S.
Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs
Matthies D, Bae C, Toombes G, Fox T, Bartesaghi A, Subramaniam S, Swartz K.
Atomic Resolution Cryo-EM Structure of β-Galactosidase
Bartesaghi A, Aguerrebere C, Falconieri V, Banerjee S, Earl LA, Zhu X, Grigorieff N, Milne JLS, Sapiro G, Wu X, Subramaniam S.
Microbiology catches the cryo-EM bug
Earl L, Falconieri V, Subramaniam S.
Cryo-EM structure of human rhodopsin bound to an inhibitory G protein
Kang Y, Kuybeda O, de Waal PW, Mukherjee S, Van Eps N, Dutka P, Zhou XE, Bartesaghi A, Erramilli S, Morizumi T, Gu X, Yin Y, Liu P, Jiang Y, Meng X, Zhao G, Melcher K, Ernst OP, Kossiakoff AA, Subramaniam S, Xu HE.
Deep-learning-assisted Volume Visualization
Cheng H-C, Cardone A, Jain S, Krokos E, Narayan K, Subramaniam S, Varshney A.
IEEE Transactions on Visualization and Computer Graphics 25: 1378-91 (2018)
Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N
Chittori S, Hong J, Saunders H, Feng H, Ghirlando R, Kelly AE, Bai Y, Subramaniam S.
Cryo-EM structures reveal mechanism and inhibition of DNA targeting by a CRISPR-Cas surveillance complex
Guo TW, Bartesaghi A, Yang H, Falconieri V, Rao P, Merk A, Eng ET, Raczkowski AM, Fox T, Earl LA, Patel D, Subramaniam S.
Broadly protective murine monoclonal antibodies against influenza B virus target highly conserved neuraminidase epitopes
Wohlbold TJ, Podolsky KA, Chromikova V, Kirkpatrick E, Falconieri V, Meade P, Amanat F, Tan J, tenOever BR, Tan GS, Subramaniam S, Palese P, Krammer F.
Building bridges between cellular and molecular structural biology
Patwardhan A, Brandt R, Butcher SJ, Collinson L, Gault D, Grünewald K, Hecksel C, Huiskonen JT, Iudin A, Jones ML, Korir PK, Koster AJ, Lagerstedt I, Lawson CL, Mastronarde D, McCormick M, Parkinson H, Rosenthal PB, Saalfeld S, Saibil HR, Sarntivijai S, Solanes Valero I, Subramaniam S, Swedlow JR, Tudose I, Winn M, Kleywegt GJ.
Cryo-EM: beyond the microscope
Earl LA, Falconieri V, Milne JLS, Subramaniam S.
Power Grid Protection of the Muscle Mitochondrial Reticulum
Glancy B, Hartnell LM, Combs CA, Fenmou A, Sun J, Murphy E, Subramaniam S, Balaban RS.
Structural basis of kainate subtype glutamate receptor desensitization
Meyerson JR, Chittori S, Merk A, Rao P, Han TH, Serpe M, Mayer ML, Subramaniam S.
Structural basis for early-onset neurological disorders caused by mutations in human selenocysteine synthase
Puppala AK, French RL, Matthies D, Baxa U, Subramaniam S, Simonović M.
Resolution advances in cryo-EM enable application to drug discovery
Subramaniam S, Earl LA, Falconieri V, Milne JLS, Egelman EH.
Mapping of Ebolavirus Neutralization by Monoclonal Antibodies in the ZMapp Cocktail Using Cryo-Electron Tomography and Studies of Cellular Entry
Tran EE, Nelson EA, Bonagiri P, Simmons JA, Shoemaker CJ, Schmaljohn CS, Kobinger GP, Zeitlin L, Subramaniam S, White JM.
The democratization of cryo-EM
Stuart DI, Subramaniam S, Abrescia NGA.
Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery
Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis M, Pragani R, Boxer M, Earl LA, Milne JLS, Subramaniam S.
Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle
Frank GA, Shukla S, Rao P, Borgnia MJ, Bartesaghi A, Merk A, Mobin A, Esser L, Earl LA, Gottesman MM, Xia D, Ambudkar SV, Subramaniam S.
Using cryo-EM to map small ligands on dynamic metabolic enzymes: studies with glutamate dehydrogenase
Borgnia MJ, Banerjee S, Merk A, Matthies D, Bartesaghi A, Rao P, Pierson J, Earl LA, Falconieri V, Subramaniam S, Milne JLS.
Cryo-electron microscopy structures of chimeric hemagglutinin displayed on a universal influenza vaccine candidate
Tran EEH, Podolsky KA, Bartesaghi A, Kuybeda O, Grandinetti G, Wohlbold TJ, Tan GS, Nachbagauer R, Palese P, Krammer F, Subramaniam S.
Cryo-EM structures of the magnesium channel CorA reveal symmetry break upon gating
Matthies D, Dalmas O, Borgnia MJ, Dominik PK, Merk A, Rao P, Reddy BG, Islam S, Bartesaghi A, Perozo E, Subramaniam S.
The crystal structure of human GlnRS provides basis for the development of neurological disorders
Ognjenović J, Wu J, Matthies D, Baxa U, Subramaniam S, Ling J, Simonović M.
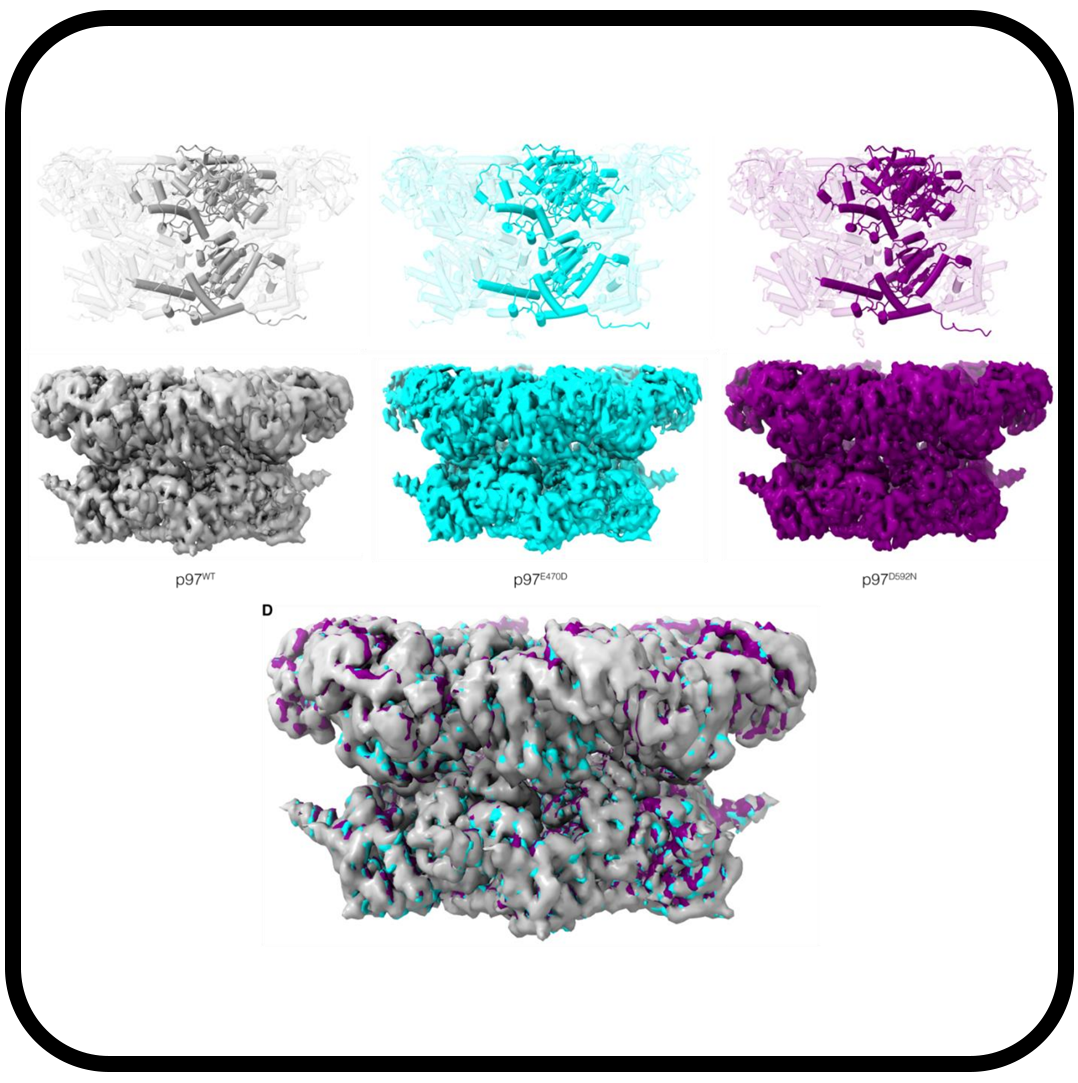
2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition
Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JLS, Huryn D, Arkin M, Subramaniam S.
Imaging cellular architecture with 3D SEM
Hartnell LM, Earl LA, Bliss D, Moran A, Subramaniam S.
Mitochondrial reticulum for cellular energy distribution in muscle
Glancy B, Hartnell LM, Malide D, Yu Z, Combs CA, Connelly PS, Subramaniam S, Balaban RS.
Cryo-EM structure of the bacteriophage T4 portal protein assembly at near-atomic resolution
Sun L, Zhang X, Gao S, Rao PA, Padilla-Sanchez V, Chen Z, Sun S, Xiang Y, Subramaniam S, Rao VB, Rossmann MG.
2.2 Å resolution cryo-EM structure of β-galactosidase in complex with a cell-permeant inhibitor
Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne J, Subramaniam S.
A versatile nano display platform from bacterial spore coat proteins
Wu IL, Narayan K, Castaing JP, Tian F, Subramaniam S, Ramamurthi KS.
Griffithsin tandemers: flexible and potent lectin inhibitors of the human immunodeficiency virus
Moulaei T, Alexandre KB, Shenoy SR, Meyerson J, Krumpe LR, Constantine B, Wilson J, Buckheit R, McMahon JB, Subramaniam S, Wlodawer A, O’Keefe BR.
Maturation of the HIV-1 core by a non-diffusional phase transition
Frank GA, Narayan K, Bess JW, Del Prete GQ, Wu X, Moran A, Hartnell LM, Earl LA, Lifson JD, Subramaniam S.
Self-assembled monolayers improve protein distribution on holey carbon cryo-EM supports
Meyerson JR, Rao P, Kumar J, Chittori S, Banerjee S, Pierson J, Mayer ML, Subramaniam S.
A 3D cellular context for the macromolecular world
Patwardhan A, Ashton A, Brandt R, Butcher S, Carzaniga R, Chiu W, Collinson L, Doux P, Duke E, Ellisman MH, Franken E, Grunewald K, Heriche JK, Koster A, Kuhlbrandt W, Lagerstedt I, Larabell C, Lawson CL, Saibil HR, Sanz-Garcia E, Subramaniam S, Verkade P, Swedlow JR, Kleywegt GJ.
Three-dimensional imaging of viral infections
Risco C, de Castro IF, Sanz-Sanchez L, Narayan K, Grandinetti G, Subramaniam S.
Structural mechanism of glutamate receptor activation and desensitization
Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S.
Structure of β-galactosidase at 3.2-Å resolution obtained by cryo-electron microscopy
Bartesaghi A, Matthies D, Banerjee S, Merk A, Subramaniam S.
Proceedings of the National Academy of Sciences USA 111: 11709-14 (2014)
Single particle cryo-electron microscopy: progress, challenges, and perspectives for further improvement
Agard D, Cheng Y, Glaeser RM, Subramaniam S.
Spatial localization of the Ebola glycoprotein mucin-like domain using cryo-electron tomography
Tran EEH, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S.
3D imaging of HIV-1 virological synapses reveals membrane architectures involved in virus transmission
Do T, Murphy G, Earl LA, Del Prete GQ, Grandinetti G, Li GH, Estes JD, Rao P, Trubey CM, Thomas J, Spector J, Bliss D, Nath A, Lifson JD, Subramaniam S.
Multi-resolution correlative focused ion beam scanning electron microscopy: Applications to cell biology
Narayan K, Danielson CM, Lagarec K, Lowekamp BC, Coffman P, Laquerre A, Phaneuf MW, Hope TJ, Subramaniam S.
Single-Particle Cryo-Electron Microscopy (Cryo-EM): Progress, Challenges, and Perspectives for Further Improvement
Agard, D, Cheng YF, Glaeser RM, Subramaniam S
Advances in Imaging and Electron Physics, Vol 185 2014, 185:113-137.
Accelerating Discovery in 3D Microanalysis: Leveraging Open Source Software and Deskside High Performance Computing
Yoo, TS, Lowekamp BC, Kuybeda O, Narayan K, Frank GA, Bartesaghi A, Borgnia M, Subramaniam S, Sapiro G, Ackerman MJ
Microscopy and Microanalysis 2014, 20:774-775.
Structure of Beta-Galactosidase at 3.2 Å Resolution Obtained by Cryo-Electron Microscopy
Bartesaghi, A, Matthies D, Banerjee S, Merk A, Subramaniam S
Proc Natl Acad Sci U S A 2014, 111:11709-11714.
Multi-Resolution Correlative Focused Ion Beam Scanning Electron Microscopy: Applications to Cell Biology
Narayan, K, Danielson CM, Lagarec K, Lowekamp BC, Coffman P, Laquerre A, Phaneuf MW, Hope TJ, Subramaniam S
J Struct Biol 2014, 185:278-284.
Three-Dimensional Imaging of Viral Infections
Risco, C, de Castro IF, Sanz-Sanchez L, Narayan K, Grandinetti G, Subramaniam S
Annu Rev Virol 2014, 1:453-473.
Self-Assembled Monolayers Improve Protein Distribution on Holey Carbon Cryo-EM Supports
Meyerson, JR, Rao P, Kumar J, Chittori S, Banerjee S, Pierson J, Mayer ML, Subramaniam S
A 3D Cellular Context for the Macromolecular World
Patwardhan, A, Ashton A, Brandt R, Butcher S, Carzaniga R, Chiu W, Collinson L, Doux P, Duke E, Ellisman MH, Franken E, Grunewald K, Heriche JK, Koster A, Kuhlbrandt W, Lagerstedt I, Larabell C, Lawson CL, Saibil HR, Sanz-Garcia E, Subramaniam S, Verkade P, Swedlow JR, Kleywegt GJ
Nat Struct Mol Biol 2014, 21:841-845.
Structural Mechanism of Glutamate Receptor Activation and Desensitization
Meyerson, JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S
Three-Dimensional Imaging of Hiv-1 Virological Synapses Reveals Membrane Architectures Involved in Virus Transmission
Do, T, Murphy G, Earl LA, Del Prete GQ, Grandinetti G, Li GH, Estes JD, Rao P, Trubey CM, Thomas J, Spector J, Bliss D, Nath A, Lifson JD, Subramaniam S
Spatial Localization of the Ebola Virus Glycoprotein Mucin-Like Domain Determined by Cryo-Electron Tomography
Tran, EE, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S
Complexes of Neutralizing and Non-Neutralizing Affinity Matured fAbs with a Mimetic of the Internal Trimeric Coiled-Coil of HIV-1 gp41
Gustchina, E, Li M, Ghirlando R, Schuck P, Louis JM, Pierson J, Rao P, Subramaniam S, Gustchina A, Clore GM, Wlodawer A
Derivation of Neural Stem Cells from Human Adult Peripheral CD34+ Cells for an Autologous Model of Neuroinflammation
Wang, T, Choi E, Monaco MC, Campanac E, Medynets M, Do T, Rao P, Johnson KR, Elkahloun AG, Von Geldern G, Johnson T, Subramaniam S, Hoffman DA, Major E, Nath A
Chemotaxis Receptor Arrays
Venkatesh, MJ, Subramaniam S
Encyclopedia of Biophysics. 2013
HIV-1 Envelope Glycoprotein Structure
Merk, A, Subramaniam S
Curr Opin Struct Biol 2013, 23:268-276.
Glutamate Receptor Desensitization Is Mediated by Changes in Quaternary Structure of the Ligand Binding Domain
Schauder, DM, Kuybeda O, Zhang J, Klymko K, Bartesaghi A, Borgnia MJ, Mayer ML, Subramaniam S
Proc Natl Acad Sci U S A 2013, 110:5921-5926.
Catching HIV ‘in the Act’ with 3D Electron Microscopy
Earl, LA, Lifson JD, Subramaniam S
Trends Microbiol 2013, 21:397-404.
Prefusion Structure of Trimeric HIV-1 Envelope Glycoprotein Determined by Cryo-Electron Microscopy
Bartesaghi, A, Merk A, Borgnia MJ, Milne JL, Subramaniam S
Nat Struct Mol Biol 2013, 20:1352-1357.
A Collaborative Framework for 3D Alignment and Classification of Heterogeneous Subvolumes in Cryo-Electron Tomography
Kuybeda, O, Frank GA, Bartesaghi A, Borgnia M, Subramaniam S, Sapiro G
J Struct Biol 2013, 181:116-127.
Cryo-Electron Microscopy–a Primer for the Non-Microscopist
Milne, JL, Borgnia MJ, Bartesaghi A, Tran EE, Earl LA, Schauder DM, Lengyel J, Pierson J, Patwardhan A, Subramaniam S
Molecular Structures of Trimeric Hiv-1 Env in Complex with Small Antibody Derivatives
Meyerson, JR, Tran EE, Kuybeda O, Chen W, Dimitrov DS, Gorlani A, Verrips T, Lifson JD, Subramaniam S
Proc Natl Acad Sci U S A 2013, 110:513-518.
HIV-1 Envelope Glycoprotein Trimers Display Open Quaternary Conformation When Bound to the Gp41 Membrane-Proximal External-Region-Directed Broadly Neutralizing Antibody Z13e1
Harris, AK, Bartesaghi A, Milne JL, Subramaniam S
Structure and Accessibility of HA Trimers on Intact 2009 H1N1 Pandemic Influenza Virus to Stem Region-Specific Neutralizing Antibodies
Harris, AK, Meyerson JR, Matsuoka Y, Kuybeda O, Moran A, Bliss D, Das SR, Yewdell JW, Sapiro G, Subbarao K, Subramaniam S
Proc Natl Acad Sci U S A 2013, 110:4592-4597.
Structure of Trimeric HIV-1 Envelope Glycoproteins
Subramaniam, S
Proc Natl Acad Sci U S A 2013, 110:E4172-4174.
Targeted Conformational Search with Map-Restrained Self-Guided Langevin Dynamics: Application to Flexible Fitting into Electron Microscopic Density Maps
Wu, X, Subramaniam S, Case DA, Wu KW, Brooks BR
J Struct Biol 2013, 183:429-440.
There is No Overkill in Biochemistry
Karnik, S, Subramaniam S
Structural Mechanism of Trimeric HIV-1 Envelope Glycoprotein Activation
Tran, EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S
Shape-Based Regularization of Electron Tomographic Reconstruction
Gopinath, A, Xu G, Ress D, Oktem O, Subramaniam S, Bajaj C
IEEE Trans Med Imaging 2012, 31:2241-2252.
Data Management Challenges in Three-Dimensional EM
Patwardhan, A, Carazo JM, Carragher B, Henderson R, Heymann JB, Hill E, Jensen GJ, Lagerstedt I, Lawson CL, Ludtke SJ, Mastronarde D, Moore WJ, Roseman A, Rosenthal P, Sorzano CO, Sanz-Garcia E, Scheres SH, Subramaniam S, Westbrook J, Winn M, Swedlow JR, Kleywegt GJ
Nat Struct Mol Biol 2012, 19:1203-1207.
Protein Secondary Structure Determination by Constrained Single-Particle Cryo-Electron Tomography
Bartesaghi, A, Lecumberry F, Sapiro G, Subramaniam S
Computational Separation of Conformational Heterogeneity Using Cryo-Electron Tomography and 3D Sub-Volume Averaging
Frank, GA, Bartesaghi A, Kuybeda O, Borgnia MJ, White TA, Sapiro G, Subramaniam S
J Struct Biol 2012, 178:165-176.
Chemical Mapping of Mammalian Cells by Atom Probe Tomography
Narayan, K, Prosa TJ, Fu J, Kelly TF, Subramaniam S
J Struct Biol 2012, 178:98-107.
Visualizing Cells and Humans in 3D: Biomedical Image Analysis at Nanometer and Meter Scales
Yoo, TS, Bliss D, Lowekamp BC, Chen DT, Murphy GE, Narayan K, Hartnell LM, Do T, Subramaniam S
IEEE Comput Graph Appl 2012, 32:39-49.
Correlative 3D Imaging of Whole Mammalian Cells with Light and Electron Microscopy
Murphy, GE, Narayan K, Lowekamp BC, Hartnell LM, Heymann JA, Fu J, Subramaniam S
J Struct Biol 2011, 176:268-278.
Three-Dimensional Structures of Soluble CD4-Bound States of Trimeric Simian Immunodeficiency Virus Envelope Glycoproteins Determined by Using Cryo-Electron Tomography
White, TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S
Determination of Molecular Structures of HIV Envelope Glycoproteins Using Cryo-Electron Tomography and Automated Sub-Tomogram Averaging
Meyerson, JR, White TA, Bliss D, Moran A, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Schauder D, Hartnell LM, Nandwani R, Dawood M, Kim B, Kim JH, Sununu J, Yang L, Bhatia S, Subramaniam C, Hurt DE, Gaudreault L, Subramaniam S
J Vis Exp 2011, 10.3791/2770:e2770.
Chemotaxis Kinase Chea Is Activated by Three Neighbouring Chemoreceptor Dimers as Effectively as by Receptor Clusters
Li, M, Khursigara CM, Subramaniam S, Hazelbauer GL
Mol Microbiol 2011, 79:677-685.
Compositional Mapping of the Surface and Interior of Mammalian Cells at Submicrometer Resolution
Szakal, C, Narayan K, Fu J, Lefman J, Subramaniam S
Trimeric HIV-1 Glycoprotein gp140 Immunogens and Native HIV-1 Envelope Glycoproteins Display the Same Closed and Open Quaternary Molecular Architectures
Harris, A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, Klasse PJ, Milne JL, Wilson IA, Olson WC, Moore JP, Subramaniam S
Proc Natl Acad Sci U S A 2011, 108:11440-11445.
Structural Plasticity of a Transmembrane Peptide Allows Self-Assembly into Biologically Active Nanoparticles
Tarasov, SG, Gaponenko V, Howard OM, Chen Y, Oppenheim JJ, Dyba MA, Subramaniam S, Lee Y, Michejda C, Tarasova NI
Proc Natl Acad Sci U S A 2011, 108:9798-9803.
Spiral Architecture of the Nucleoid in Bdellovibrio Bacteriovorus
Butan, C, Hartnell LM, Fenton AK, Bliss D, Sockett RE, Subramaniam S, Milne JL
J Bacteriol 2011, 193:1341-1350.
Lateral Density of Receptor Arrays in the Membrane Plane Influences Sensitivity of the E. Coli Chemotaxis Response
Khursigara, CM, Lan G, Neumann S, Wu X, Ravindran S, Borgnia MJ, Sourjik V, Milne J, Tu Y, Subramaniam S
HIV-1 Activates CDC42 and Induces Membrane Extensions in Immature Dendritic Cells to Facilitate Cell-to-Cell Virus Propagation
Nikolic, DS, Lehmann M, Felts R, Garcia E, Blanchet FP, Subramaniam S, Piguet V
A Coiled-Coil-Repeat Protein ‘Ccrp’ in Bdellovibrio Bacteriovorus Prevents Cellular Indentation, but Is Not Essential for Vibroid Cell Morphology
Fenton, AK, Hobley L, Butan C, Subramaniam S, Sockett RE
FEMS Microbiology Letters 2010, 313:89-95.
Ion-Abrasion Scanning Electron Microscopy Reveals Distorted Liver Mitochondrial Morphology in Murine Methylmalonic Acidemia
Murphy, GE, Lowekamp BC, Zerfas PM, Chandler RJ, Narasimha R, Venditti CP, Subramaniam S
J Struct Biol 2010, 171:125-132.
Molecular Architectures of Trimeric SIV and HIV-1 Envelope Glycoproteins on Intact Viruses: Strain-Dependent Variation in Quaternary Structure
White, TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S
3D Visualization of HIV Transfer at the Virological Synapse between Dendritic Cells and T Cells
Felts, RL, Narayan K, Estes JD, Shi D, Trubey CM, Fu J, Hartnell LM, Ruthel GT, Schneider DK, Nagashima K, Bess JW, Jr., Bavari S, Lowekamp BC, Bliss D, Lifson JD, Subramaniam S
Proc Natl Acad Sci U S A 2010, 107:13336-13341.
3D Imaging of Mammalian Cells with Ion-Abrasion Scanning Electron Microscopy
Heymann, JA, Shi D, Kim S, Bliss D, Milne JL, Subramaniam S
Membrane Protein Structure Determination Using Cryo-Electron Tomography and 3D Image Averaging
Bartesaghi, A, Subramaniam S
Curr Opin Struct Biol 2009, 19:402-407.
Structural Snapshots of Conformational Changes in a Seven-Helix Membrane Protein: Lessons from Bacteriorhodopsin
Hirai, T, Subramaniam S, Lanyi JK
Curr Opin Struct Biol 2009, 19:433-439.
Staphylococcal Enterotoxin a Induces Small Clusters of HLA-DR1 on B Cells
Narayan, K, Perkins EM, Murphy GE, Dalai SK, Edidin M, Subramaniam S, Sadegh-Nasseri S
3D Imaging of Diatoms with Ion-Abrasion Scanning Electron Microscopy
Hildebrand, M, Kim S, Shi D, Scott K, Subramaniam S
J Struct Biol 2009, 166:316-328.
Automatic Joint Classification and Segmentation of Whole Cell 3D Images
Narasimha, R, Ouyang H, Gray A, McLaughlin SW, Subramaniam S
Pattern Recognition 2009, 42:1067-1079.
Protein Conformational Changes in the Bacteriorhodopsin Photocycle: Comparison of Findings from Electron and X-Ray Crystallographic Analyses
Hirai, T, Subramaniam S
Ion-Abrasion Scanning Electron Microscopy Reveals Surface-Connected Tubular Conduits in HIV-Infected Macrophages
Bennett, AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S
Cryo-Electron Tomography of Bacteria: Progress, Challenges and Future Prospects
Milne, JL, Subramaniam S
Nat Rev Microbiol 2009, 7:666-675.
3D Imaging of Diatoms with Ion Abrasion Scanning Electron Microscopy
Hildebrand, M, Kim S, Shi D, Subramaniam S
Microscopy and Microanalysis 2008, 14:972-973.
Molecular Architecture of Native HIV-1 gp 120 Trimers
Liu, J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S
Three-Dimensional Imaging of the Highly Bent Architecture of Bdellovibrio Bacteriovorus by Using Cryo-Electron Tomography
Borgnia, MJ, Subramaniam S, Milne JL
J Bacteriol 2008, 190:2588-2596.
Electron Tomography in Nanoparticle Imaging and Analysis
Lengyel, JS, Milne JL, Subramaniam S
Nanomedicine (Lond) 2008, 3:125-131.
Stoichiometry and Absolute Quantification of Proteins with Mass Spectrometry Using Fluorescent and Isotope-Labeled Concatenated Peptide Standards
Nanavati, D, Gucek M, Milne JL, Subramaniam S, Markey SP
Mol Cell Proteomics 2008, 7:442-447.
Extended Polypeptide Linkers Establish the Spatial Architecture of a Pyruvate Dehydrogenase Multienzyme Complex
Lengyel, JS, Stott KM, Wu X, Brooks BR, Balbo A, Schuck P, Perham RN, Subramaniam S, Milne JL
Classification and 3D Averaging with Missing Wedge Correction in Biological Electron Tomography
Bartesaghi, A, Sprechmann P, Liu J, Randall G, Sapiro G, Subramaniam S
J Struct Biol 2008, 162:436-450.
Chemoreceptors in Caulobacter Crescentus: Trimers of Receptor Dimers in a Partially Ordered Hexagonally Packed Array
Khursigara, CM, Wu X, Subramaniam S
J Bacteriol 2008, 190:6805-6810.
Evaluation of Denoising Algorithms for Biological Electron Tomography
Narasimha, R, Aganj I, Bennett AE, Borgnia MJ, Zabransky D, Sapiro G, McLaughlin SW, Milne JL, Subramaniam S
Role of HAMP Domains in Chemotaxis Signaling by Bacterial Chemoreceptors
Khursigara, CM, Wu X, Zhang P, Lefman J, Subramaniam S
Proc Natl Acad Sci U S A 2008, 105:16555-16560.
Regularization for Inverting the Radon Transform with Wedge Consideration
Aganj, L, Bartesaghi A, Borgnia M, Liao HY, Sapiro G, Subramaniam S
2007 4th IEEE International Symposium on Biomedical Imaging : Macro to Nano, Vols 1-3 2007
Classification, Averaging and Reconstruction of Macromolecules in Electron Tomography
Bartesaghi, A, Sprechmann P, Randall G, Sapiro G, Subramaniam S
2007 4th IEEE International Symposium on Biomedical Imaging : Macro to Nano, Vols 1-3 2007,
From Gigabytes to Bytes: Automated Denoising and Feature Identification in Electron Tomograms of Intact Bacterial Cells
Narasimha, R, Aganj I, Borgnia M, Sapiro G, McLaughlin S, Milne J, Subramaniamo S
2007 4th IEEE International Symposium on Biomedical Imaging : Macro to Nano, Vols 1-3 2007
Gigabytes to Bytes: Automated Denoising and Feature Extraction as Applied to the Analysis of HIV Architecture and Variability Using Electron Tomography
Narasimha, R, Bennett A, Zabransky D, Sougrat R, McLaughlin S, Subramaniam S
Determination of Protein Structures in Situ: Electron Tomography of Intact Viruses and Cells
Subramaniam, S
2007 4th IEEE International Symposium on Biomedical Imaging : Macro to Nano, Vols 1-3 2007
Electron Tomography of Bacterial Chemotaxis Receptor Assemblies
Zhang, P, Weis RM, Peters PJ, Subramaniam S
Methods Cell Biol 2007, 79:373-384.
Automated 100-Position Specimen Loader and Image Acquisition System for Transmission Electron Microscopy
Lefman, J, Morrison R, Subramaniam S
J Struct Biol 2007, 158:318-326.
Direct Visualization of Escherichia Coli Chemotaxis Receptor Arrays Using Cryo-Electron Microscopy
Zhang, P, Khursigara CM, Hartnell LM, Subramaniam S
Proc Natl Acad Sci U S A 2007, 104:3777-3781.
Electron Tomography of the Contact between T Cells and SIV/HIV-1: Implications for Viral Entry
Sougrat, R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S
Cell Surface Filaments of the Gliding Bacterium Flavobacterium Johnsoniae Revealed by Cryo-Electron Tomography
Liu, J, McBride MJ, Subramaniam S
J Bacteriol 2007, 189:7503-7506.
Electron Tomography of Viruses
Subramaniam, S, Bartesaghi A, Liu J, Bennett AE, Sougrat R
Curr Opin Struct Biol 2007, 17:596-602.
Cryoelectron Tomographic Analysis of an HIV-Neutralizing Protein and Its Complex with Native Viral gp120
Bennett, A, Liu J, Van Ryk D, Bliss D, Arthos J, Henderson RM, Subramaniam S
J Biol Chem 2007, 282:27754-27759.
Automated, 100-Specimen Loading and Imaging System for Transmission Electron Microscopy
Lefman, J, Morrison R, Subramaniam S
Microscopy and Microanalysis 2006, 12:1100-1101.
Structure and Function of Icosahedral Pyruvate Dehydrogenase Complexes
Milne J, Wu X, Borgnia M, Lengyel J, Shi D, Perham R, Subramaniam S
Microscopy and Microanalysis. Cambridge University Press; 2006;12(S02):26–27.
Bridging the Imaging Gap in Nanobiology with Three-Dimensional Electron Microscopy
Subramaniam, S
2006 Bio- Micro- and Nanosystems Conference 2006
The SIV Surface Spike Imaged by Electron Tomography: One Leg or Three?
Subramaniam, S
Molecular Structure of a 9-MDa Icosahedral Pyruvate Dehydrogenase Subcomplex Containing the E2 and E3 Enzymes Using Cryoelectron Microscopy
Milne, JL, Wu X, Borgnia MJ, Lengyel JS, Brooks BR, Shi D, Perham RN, Subramaniam S
J Biol Chem 2006, 281:4364-4370.
Site-Specific 3D Imaging of Cells and Tissues with a Dual Beam Microscope
Heymann, JA, Hayles M, Gestmann I, Giannuzzi LA, Lich B, Subramaniam S
J Struct Biol 2006, 155:63-73.
Bridging the Imaging Gap: Visualizing Subcellular Architecture with Electron Tomography
Subramaniam, S
Curr Opin Microbiol 2005, 8:316-322.
Electron Tomography of Degenerating Neurons in Mice with Abnormal Regulation of Iron Metabolism
Zhang, P, Land W, Lee S, Juliani J, Lefman J, Smith SR, Germain D, Kessel M, Leapman R, Rouault TA, Subramaniam S
J Struct Biol 2005, 150:144-153.
An Energy-Based Three-Dimensional Segmentation Approach for the Quantitative Interpretation of Electron Tomograms
Bartesaghi, A, Sapiro G, Subramaniam S
IEEE Trans Image Process 2005, 14:1314-1323.
A New Approach for 3D Segmentation of Cellular Tomograms Obtained Using Three-Dimensional Electron Microscopy
Bartesaghi, A, Sapiro G, Lee S, Lefman J, Wahl S, Orenstein J, Subramaniam S
Site-Specific 3D Imaging of Cells and Tissues Using Dualbeam Technology
Gestmann, I, Hayles M, Shi D, Kumar G, Giannuzzi L, Lich B, Subramaniam S
Microscopy and Microanalysis 2004, 10:1124-1125.
Structure and Transport Mechanism of the Bacterial Oxalate Transporter OxIT
Hirai, T, Subramaniam S
Biophysical Journal 2004, 87:3600-3607.
A Template-Propagation Method for Segmentation of Filamentous Structures in Electron Tomograms
Shen, WC, Zhang PJ, Germain D, Rouault T, Subramaniam S
Three-Dimensional Electron Microscopy at Molecular Resolution
Subramaniam, S, Milne JL
Annu Rev Biophys Biomol Struct 2004, 33:141-155.
Three-Dimensional Electron Microscopic Imaging of Membrane Invaginations in Escherichia Coli Overproducing the Chemotaxis Receptor Tsr
Lefman, J, Zhang P, Hirai T, Weis RM, Juliani J, Bliss D, Kessel M, Bos E, Peters PJ, Subramaniam S
J Bacteriol 2004, 186:5052-5061.
Direct Visualization of Receptor Arrays in Frozen-Hydrated Sections and Plunge-Frozen Specimens of E. Coli Engineered to Overproduce the Chemotaxis Receptor Tsr
Zhang, P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S
3D Imaging of Membrane Networks in a Bacterial Cell Using Electron Tomography
Zhang, P, Lefman J, Kessel M, Juliani J, Hirai T, Weis RM, Peters P, Subramaniam S
Microscopy and Microanalysis 2003, 9:1168-1169.
Automated Image Acquisition and Processing Using a New Generation of 4k×4k CCD Cameras for Cryo Electron Microscopic Studies of Macromolecular Assemblies
Zhang, P, Borgnia MJ, Mooney P, Shi D, Pan M, O’Herron P, Mao A, Brogan D, Milne JLS, Subramaniam S
Journal of Structural Biology 2003, 143:135-144.
Projection Structure of the Bacterial Oxalate Transporter Oxlt at 3.4 Å Resolution
Heymann, JA, Hirai T, Shi D, Subramaniam S
J Struct Biol 2003, 144:320-326.
A Core-Weighted Fitting Method for Docking Atomic Structures into Low-Resolution Maps: Application to Cryo-Electron Microscopy
Wu, X, Milne JLS, Borgnia MJ, Rostapshov AV, Subramaniam S, Brooks BR
Journal of Structural Biology 2003, 141:63-76.
Electron Microscopic Analysis of Membrane Assemblies Formed by the Bacterial Chemotaxis Receptor Tsr
Weis, RM, Hirai T, Chalah A, Kessel M, Peters PJ, Subramaniam S
J Bacteriol 2003, 185:3636-3643.
Structural Insights into the Mechanism of Proton Pumping by Bacteriorhodopsin
Hirai, T, Subramaniam S
Structural Model for 12-Helix Transporters Belonging to the Major Facilitator Superfamily
Hirai, T, Heymann JA, Maloney PC, Subramaniam S
J Bacteriol 2003, 185:1712-1718.
Molecular Structure of an Icosahedral Pyruvate Dehydrogenase Complex
Subramaniam, S, Shi D, Perham RN, Milne JLS
Microscopy and Microanalysis 2002, 8:214-215.
Three-Dimensional Structure of a Bacterial Oxalate Transporter
Hirai, T, Heymann JA, Shi D, Sarker R, Maloney PC, Subramaniam S
Nat Struct Biol 2002, 9:597-600.
From Structure to Mechanism: Electron Crystallographic Studies of Bacteriorhodopsin
Subramaniam, S, Hirai T, Henderson R
Philos Trans A Math Phys Eng Sci 2002, 360:859-874.
Proton Translocation by Bacteriorhodopsin in the Absence of Substantial Conformational Changes
Tittor, J, Paula S, Subramaniam S, Heberle J, Henderson R, Oesterhelt D
Molecular Architecture and Mechanism of an Icosahedral Pyruvate Dehydrogenase Complex: A Multifunctional Catalytic Machine
Milne, JL, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, Brooks BR, Perham RN, Henderson R, Subramaniam S
Projection Structure and Molecular Architecture of Oxlt, a Bacterial Membrane Transporter
Heymann, JA, Sarker R, Hirai T, Shi D, Milne JL, Maloney PC, Subramaniam S
Automated Data Collection with a Tecnai 12 Electron Microscope: Applications for Molecular Imaging by Cryomicroscopy
Zhang, P, Beatty A, Milne JL, Subramaniam S
J Struct Biol 2001, 135:251-261.
Crystallographic Analysis of Protein Conformational Changes in the Bacteriorhodopsin Photocycle
Subramaniam, S, Henderson R
Biochim Biophys Acta 2000, 1460:157-165.
CCD Detectors in High-Resolution Biological Electron Microscopy
Faruqi, AR, Subramaniam S
Integration of Deletion Mutants of Bovine Rhodopsin into the Membrane of the Endoplasmic Reticulum
Heymann, JAW, Subramaniam S
Molecular Membrane Biology 2000, 17:165-174.
The Structure of Bacteriorhodopsin: An Emerging Consensus
Subramaniam, S
Curr Opin Struct Biol 1999, 9:462-468.
Electron Crystallography of Bacteriorhodopsin with Millisecond Time Resolution
Subramaniam, S, Henderson R
J Struct Biol 1999, 128:19-25.
Cooled CCD Detector with Tapered Fibre Optics for Recording Electron Diffraction Patterns
Faruqi, AR, Henderson R, Subramaniam S
Ultramicroscopy 1999, 75:235-250.
Protein Conformational Changes in the Bacteriorhodopsin Photocycle
Subramaniam, S, Lindahl M, Bullough P, Faruqi AR, Tittor J, Oesterhelt D, Brown L, Lanyi J, Henderson R
Evidence for a Long-Lived 13-Cis-Containing Intermediate in the Photocycle of the Leu 93 ->Ala Bacteriorhodopsin Mutant
Delaney, JK, Schmidt P, Atkinson GH, Subramaniam S
Journal of Physical Chemistry B 1997, 101.
Expression of Bacteriorhodopsin in Sf9 and Cos-1 Cells
Heymann, J, Jager R, Subramaniam S
J Bioenerg Biomembr 1997, 29:55-59.
Studies of Rh1 Metarhodopsin Stabilization in Wild-Type Drosophila and in Mutants Lacking One or Both Arrestins
Kiselev, A, Subramaniam S
Biochemistry 1997, 36:2188-2196.
Structural Changes Underlying Proton Pumping by Bacteriorhodopsin
Henderson, R, Bullough P, Lindahl M, Subramaniam S
FASEB Journal 1997, 11:A992-A992.
Electron Diffraction Studies of Light-Induced Conformational Changes in the Leu-93 to Ala Bacteriorhodopsin Mutant
Subramaniam, S, Faruqi AR, Oesterhelt D, Henderson R
Proc Natl Acad Sci U S A 1997, 94:1767-1772.
Reducing the Flexibility of Retinal Restores a Wild-Type-Like Photocycle in Bacteriorhodopsin Mutants Defective in Protein-Retinal Coupling
Delaney, JK, Yahalom G, Sheves M, Subramaniam S
Proc Natl Acad Sci U S A 1997, 94:5028-5033.
Expression, Stability, and Membrane Integration of Truncation Mutants of Bovine Rhodopsin
Heymann, JA, Subramaniam S
Proc Natl Acad Sci U S A 1997, 94:4966-4971.
The Residues Leu 93 and Asp 96 Act Independently in the Bacteriorhodopsin Photocycle: Studies with the Leu 93-to Ala, Asp 96 to Asn Double Mutant
Delaney, JK, Subramaniam S
Biophysical Journal 1996, 70:2366-2372.
Modulation of Arrestin Release in the Light-Driven Regeneration of Rh1 Drosophila Rhodopsin
Kiselev, A, Subramaniam S
Biochemistry 1996, 35:1848-1855.
Molecular Mechanism of Protein-Retinal Coupling in Bacteriorhodopsin
Delaney, JK, Schweiger U, Subramaniam S
Proc Natl Acad Sci U S A 1995, 92:11120-11124.
Activation and Regeneration of Rhodopsin in the Insect Visual Cycle
Kiselev, A, Subramaniam S
Electron Diffraction Analysis of Structural Changes in the Photocycle of Bacteriorhodopsin
Subramaniam, S, Gerstein M, Oesterhelt D, Henderson R
Hydrophobic Amino Acids in the Retinal-Binding Pocket of Bacteriorhodopsin
Greenhalgh, DA, Farrens DL, Subramaniam S, Khorana HG
J Biol Chem 1993, 268:20305-20311.
Effect of Introducing Different Carboxylate-Containing Side-Chains at Position-85 on Chromophore Formation and Proton Transport in Bacteriorhodopsin
Greenhalgh, DA, Subramaniam S, Alexiev U, Otto H, Heyn MP, Khorana HG
Journal of Biological Chemistry 1992, 267:25734-25738.
Aspartic Acid-85 in Bacteriorhodopsin Functions Both as Proton Acceptor and Negative Counterion to the Schiff-Base
Subramaniam, S, Greenhalgh DA, Khorana HG
Journal of Biological Chemistry 1992, 267:25730-25733.
Replacement of Leucine-93 by Alanine or Threonine Slows Down the Decay of the N and O Intermediates in the Photocycle of Bacteriorhodopsin: Implications for Proton Uptake and 13-Cis-Retinal—-All-Trans-Retinal Reisomerization
Subramaniam, S, Greenhalgh DA, Rath P, Rothschild KJ, Khorana HG
Proc Natl Acad Sci U S A 1991, 88:6873-6877.
The Reaction of Hydroxylamine with Bacteriorhodopsin Studied with Mutants That Have Altered Photocycles: Selective Reactivity of Different Photointermediates
Subramaniam, S, Marti T, Rosselet SJ, Rothschild KJ, Khorana HG
Proc Natl Acad Sci U S A 1991, 88:2583-2587.
Protonation State of Asp (Glu)-85 Regulates the Purple-to-Blue Transition in Bacteriorhodopsin Mutants Arg-82—-Ala and Asp-85—-Glu: The Blue Form Is Inactive in Proton Translocation
Subramaniam, S, Marti T, Khorana HG
Proc Natl Acad Sci U S A 1990, 87:1013-1017.
Ultraviolet-Visible Transient Spectroscopy of Bacteriorhodopsin Mutants. Evidence for Two Forms of Tyrosine-185—-Phenylalanine
Dunach, M, Berkowitz S, Marti T, He YW, Subramaniam S, Khorana HG, Rothschild KJ
J Biol Chem 1990, 265:16978-16984.
Replacement of Aspartic Residues 85, 96, 115, or 212 Affects the Quantum Yield and Kinetics of Proton Release and Uptake by Bacteriorhodopsin
Marinetti, T, Subramaniam S, Mogi T, Marti T, Khorana HG
Proc Natl Acad Sci U S A 1989, 86:529-533.
Critical Mixing in Monolayer Mixtures of Phospholipid and Cholesterol
Subramaniam, S, McConnell HM
The Journal of Physical Chemistry 1987, 91:1715-1718.
Lateral Diffusion of Specific Antibodies Bound to Lipid Monolayers on Alkylated Substrates
Subramaniam, S, Seul M, McConnell HM
Proc Natl Acad Sci U S A 1986, 83:1169-1173.
Mono- and Bilayers of Phospholipids at Interfaces: Interlayer Coupling and Phase Stability
Seul, M, Subramaniam S, McConnell HM