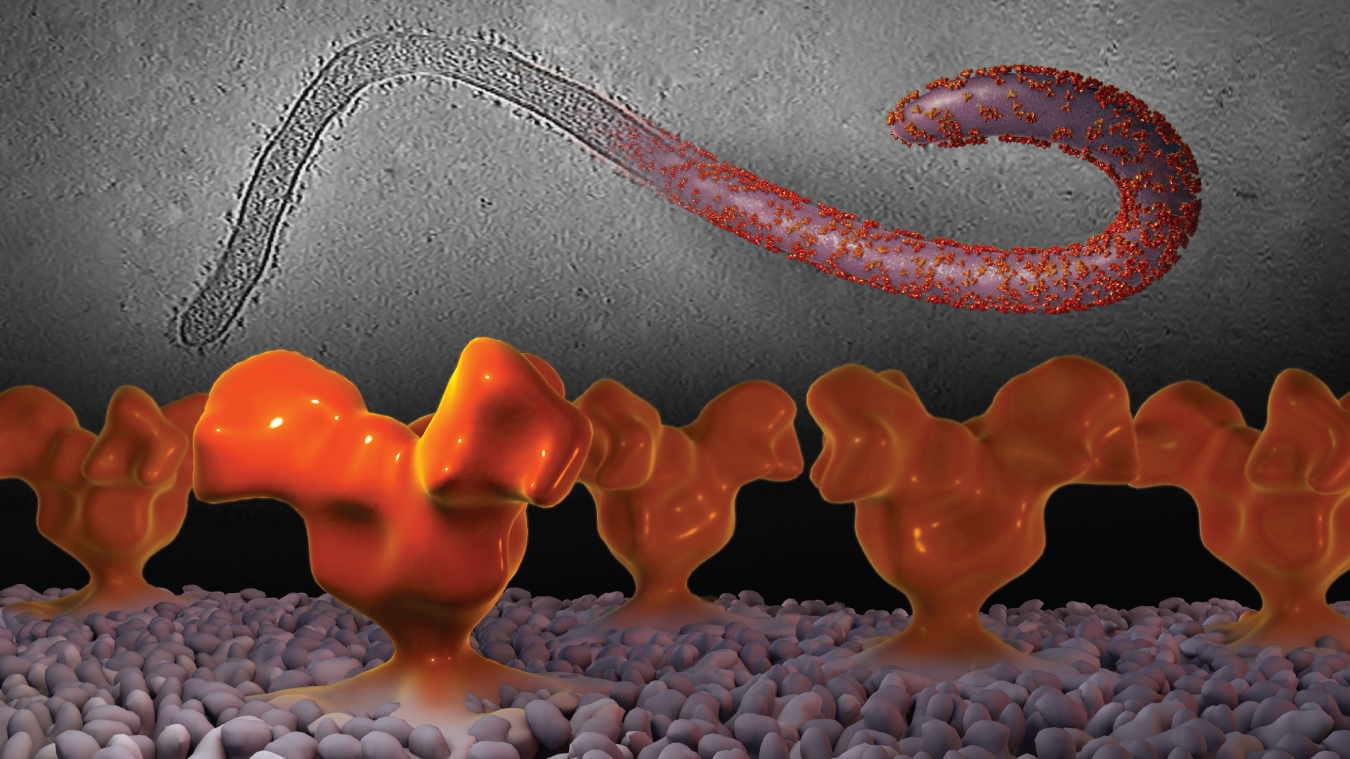
As with most viruses, the Ebola virus itself is very small – too small to be seen even in a high-powered light microscope. But when visualized in an electron microscope, the surface of the filament-shaped virus particle (also known as a virion) can be seen, studded with small proteins on its surface (upper right of the image above). It’s these proteins (shown in orange) that allow the virus to bind to and enter into cells, leading to infection.
To study the Ebola virus and the proteins on its surface, we used a of version cryo-electron tomography that we originally developed to study similar proteins in HIV. In this technique, single virions (upper left of the image above) are imaged at a variety of angles, leading to a 3D image of that virion. But then, to achieve higher resolution of the individual proteins (which are too tiny to see at great detail even by electron microscopy), the images of many of these proteins are averaged together.
The foreground of the image above shows the 3D structure of the envelope glycoprotein spike, first published in 2014, from the Zaire strain of Ebola. Like HIV, the structure of the Ebola spike has been challenging to determine, in part because a significant portion of this trimeric protein complex is composed of poorly ordered carbohydrates (sugar molecules). This was the first structure of the Ebola spike in its native state, showing that the carbohydrates are located in a cap at the apex of the spike.
Full-size image: Download (1.8MB)
Related Reference: Tran EEH, Simmons JA, et al. Spatial localization of the Ebola glycoprotein mucin-like domain using cryo-electron tomography. J Virol. 2014 Sep;88(18):10958-62. doi:10.1128/JVI00870-14.